
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,375,903 – 19,376,027 |
| Length | 124 |
| Max. P | 0.973601 |

| Location | 19,375,903 – 19,376,007 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 90.99 |
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -30.31 |
| Energy contribution | -30.53 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19375903 104 - 20766785 CAGUUGGACUUUGGCUCUGAUAAUUCAUAACGAUUUGUGGCAGCAGCAGCCGAUUGAAUCUGAAAGGGCCAAAGUCCAAACUCGA-AUUCGCAGUGACGUCUUGC ...(((((((((((((((((....)).....(((((.((((.......))))...)))))....)))))))))))))))(((((.-...)).))).......... ( -35.50) >DroSec_CAF1 2784 104 - 1 CGGUUGGACUUUGGCUCUGAUAAUUCAUAACGAUUUGUGGCAGCAGCAGCCGAUUGAAGCUGAAAGGGCCAAAGUCCAAACUCGA-AUUCGCAGUGACGUCUUGC ((.(((((((((((((((......((((((....))))))((((..(((....)))..))))..)))))))))))))))(((((.-...)).)))..))...... ( -35.60) >DroSim_CAF1 2835 104 - 1 CGGUUGGACUUUGGCUCUGAUAAUUCAUAACGAUUUGUGGCAGCAGCAGCCGAUUGAAUCUGAAAGGGCCAAAGUCCAAACUCGA-AUUCGCAGUGACGUCUUGC ((.(((((((((((((((((....)).....(((((.((((.......))))...)))))....)))))))))))))))(((((.-...)).)))..))...... ( -35.70) >DroEre_CAF1 3056 104 - 1 CGGUUGGACUUUGGCUCUGAUAAUUCAUAACGAUUUGUGGCAGCAGCAGCCGAUUGAAUCCGAAAGGGCCAAAGUCCAAACUCGA-GUUCGCAGUGACGUCUUGC ((.(((((((((((((((((....)).....(((((.((((.......))))...)))))....)))))))))))))))(((((.-...)).)))..))...... ( -35.30) >DroYak_CAF1 2840 104 - 1 CGGUUGGACUUUGGCUCUGACAAUUCAUAACGAUUUGUGGCAGCAGCAGCCGAUUGAAUCUGAAAGGGCCAAAGUCCAAACUCGA-GUUCGCAGUGACGUCUUGC ((.(((((((((((((((..(((((((..........((((.......))))..))))).))..)))))))))))))))(((((.-...)).)))..))...... ( -35.30) >DroAna_CAF1 2152 92 - 1 CGGUUGGACUUUGCCUUCAUCAAU---------UUUGUGGCAUCAGCGGCCGAUUGAAUCUCA-AGGGCCAAAGUCCAAACUCGAAAUUCGCAGUGACGUCU--- ((.((((((((((((..((.....---------..)).)))......((((..(((.....))-).)))))))))))))(((((.....)).)))..))...--- ( -28.10) >consensus CGGUUGGACUUUGGCUCUGAUAAUUCAUAACGAUUUGUGGCAGCAGCAGCCGAUUGAAUCUGAAAGGGCCAAAGUCCAAACUCGA_AUUCGCAGUGACGUCUUGC ...(((((((((((((((.((((.((.....)).))))(((.......))).............)))))))))))))))(((((.....)).))).......... (-30.31 = -30.53 + 0.22)

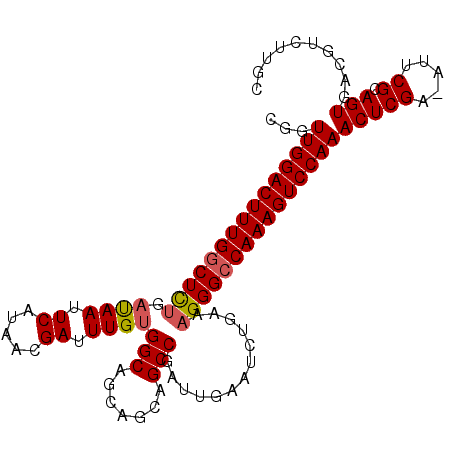
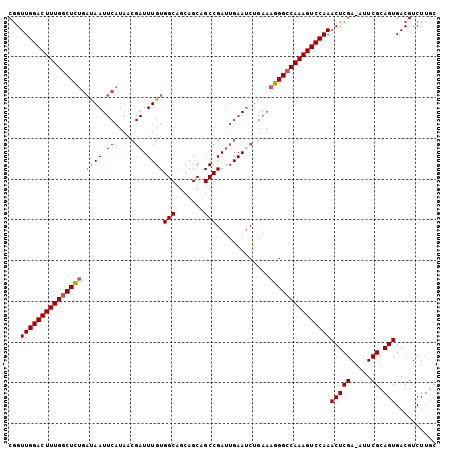
| Location | 19,375,928 – 19,376,027 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -23.74 |
| Energy contribution | -24.63 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19375928 99 + 20766785 UUGGACUUUGGCCCUUUCAGAUUCAAUCGGCUGCUGCUGCCACAAAUCGUUAUGAAUUAUCAGAGCCAAAGUCCAACUGGUG--CAGCCCGUGUUUGGAAU-- .(((((((((((.((....((((((...(((.......)))((.....))..))))))...)).)))))))))))((.((..--...)).)).........-- ( -29.70) >DroSec_CAF1 2809 99 + 1 UUGGACUUUGGCCCUUUCAGCUUCAAUCGGCUGCUGCUGCCACAAAUCGUUAUGAAUUAUCAGAGCCAAAGUCCAACCGGUG--CAGCCCGUGUCUGGAAU-- ((((((((((((.((......((((...(((.......)))((.....))..)))).....)).))))))))))))((((.(--((.....)))))))...-- ( -30.40) >DroSim_CAF1 2860 99 + 1 UUGGACUUUGGCCCUUUCAGAUUCAAUCGGCUGCUGCUGCCACAAAUCGUUAUGAAUUAUCAGAGCCAAAGUCCAACCGGUG--CAGCCCGUGUCUGGAAU-- ((((((((((((.((....((((((...(((.......)))((.....))..))))))...)).))))))))))))((((.(--((.....)))))))...-- ( -33.20) >DroEre_CAF1 3081 99 + 1 UUGGACUUUGGCCCUUUCGGAUUCAAUCGGCUGCUGCUGCCACAAAUCGUUAUGAAUUAUCAGAGCCAAAGUCCAACCGGUG--CAGCCCACGUUUAGGAU-- ((((((((((((.((....((((((...(((.......)))((.....))..))))))...)).))))))))))))((...(--(.......))...))..-- ( -31.10) >DroYak_CAF1 2865 99 + 1 UUGGACUUUGGCCCUUUCAGAUUCAAUCGGCUGCUGCUGCCACAAAUCGUUAUGAAUUGUCAGAGCCAAAGUCCAACCGGUG--CAGUAUAGCUUUGGGAU-- ((((((((((((.((....((((((...(((.......)))((.....))..))))))...)).))))))))))))((((.(--(......)).))))...-- ( -34.40) >DroAna_CAF1 2175 91 + 1 UUGGACUUUGGCCCU-UGAGAUUCAAUCGGCCGCUGAUGCCACAAA---------AUUGAUGAAGGCAAAGUCCAACCGGCAUACAGUU--AGUCAAGCAUAA (((((((((((((.(-((.....)))..))))......(((.....---------.........))))))))))))..(((........--.)))........ ( -23.34) >consensus UUGGACUUUGGCCCUUUCAGAUUCAAUCGGCUGCUGCUGCCACAAAUCGUUAUGAAUUAUCAGAGCCAAAGUCCAACCGGUG__CAGCCCGUGUCUGGAAU__ ((((((((((((.((....((((((...(((.......)))((.....))..))))))...)).))))))))))))..(((.....))).............. (-23.74 = -24.63 + 0.89)

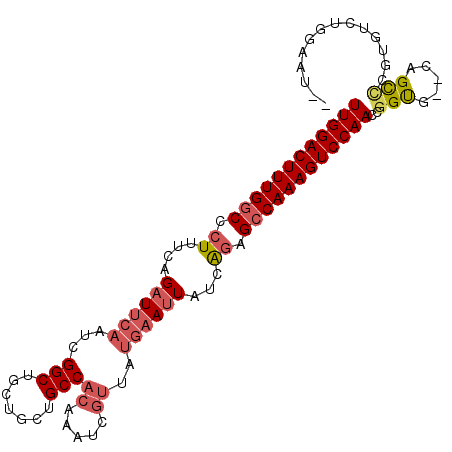
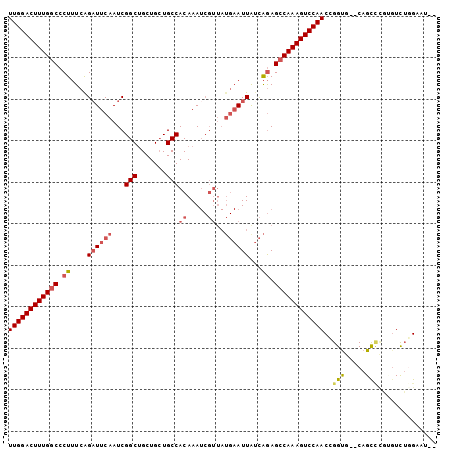
| Location | 19,375,928 – 19,376,027 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -28.00 |
| Energy contribution | -27.87 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19375928 99 - 20766785 --AUUCCAAACACGGGCUG--CACCAGUUGGACUUUGGCUCUGAUAAUUCAUAACGAUUUGUGGCAGCAGCAGCCGAUUGAAUCUGAAAGGGCCAAAGUCCAA --...........((....--..))..(((((((((((((((((....)).....(((((.((((.......))))...)))))....))))))))))))))) ( -34.70) >DroSec_CAF1 2809 99 - 1 --AUUCCAGACACGGGCUG--CACCGGUUGGACUUUGGCUCUGAUAAUUCAUAACGAUUUGUGGCAGCAGCAGCCGAUUGAAGCUGAAAGGGCCAAAGUCCAA --......(((.(((....--..))))))(((((((((((((......((((((....))))))((((..(((....)))..))))..))))))))))))).. ( -37.50) >DroSim_CAF1 2860 99 - 1 --AUUCCAGACACGGGCUG--CACCGGUUGGACUUUGGCUCUGAUAAUUCAUAACGAUUUGUGGCAGCAGCAGCCGAUUGAAUCUGAAAGGGCCAAAGUCCAA --......(((.(((....--..))))))(((((((((((((((....)).....(((((.((((.......))))...)))))....))))))))))))).. ( -37.60) >DroEre_CAF1 3081 99 - 1 --AUCCUAAACGUGGGCUG--CACCGGUUGGACUUUGGCUCUGAUAAUUCAUAACGAUUUGUGGCAGCAGCAGCCGAUUGAAUCCGAAAGGGCCAAAGUCCAA --......(((.(((....--..))))))(((((((((((((((....)).....(((((.((((.......))))...)))))....))))))))))))).. ( -34.60) >DroYak_CAF1 2865 99 - 1 --AUCCCAAAGCUAUACUG--CACCGGUUGGACUUUGGCUCUGACAAUUCAUAACGAUUUGUGGCAGCAGCAGCCGAUUGAAUCUGAAAGGGCCAAAGUCCAA --...((...((......)--)...))(((((((((((((((..(((((((..........((((.......))))..))))).))..))))))))))))))) ( -34.20) >DroAna_CAF1 2175 91 - 1 UUAUGCUUGACU--AACUGUAUGCCGGUUGGACUUUGCCUUCAUCAAU---------UUUGUGGCAUCAGCGGCCGAUUGAAUCUCA-AGGGCCAAAGUCCAA ....((...((.--....))..))...((((((((((((..((.....---------..)).)))......((((..(((.....))-).))))))))))))) ( -26.40) >consensus __AUUCCAAACACGGGCUG__CACCGGUUGGACUUUGGCUCUGAUAAUUCAUAACGAUUUGUGGCAGCAGCAGCCGAUUGAAUCUGAAAGGGCCAAAGUCCAA ...............((((.....))))((((((((((((((.((((.((.....)).))))(((.......))).............)))))))))))))). (-28.00 = -27.87 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:22 2006