| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,375,611 – 19,375,706 |
| Length | 95 |
| Max. P | 0.819668 |

| Location | 19,375,611 – 19,375,706 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -23.50 |
| Energy contribution | -24.53 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
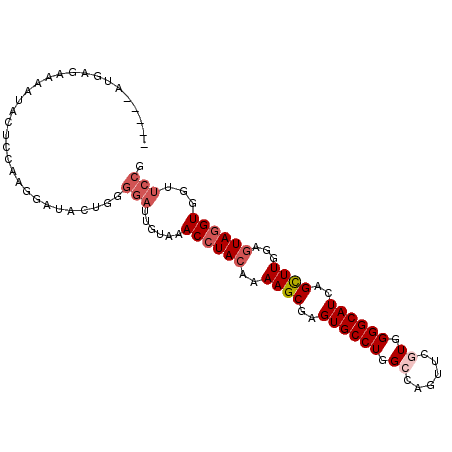
>2R_DroMel_CAF1 19375611 95 + 20766785 -----GUAAGCUAAUACUCCAAGGAUAUUGGGGAUUGUAAACCUACAAAAGCGAGUGCCUGGCCAGUUCAUGGGGCAUCAGCUUGGAGUAGGUGGUUCCG -----.....((((((.((....))))))))(((......((((((..((((..(((((...(((.....))))))))..))))...))))))...))). ( -31.70) >DroSec_CAF1 2492 95 + 1 -----GUGAGAUAAUACUCCAAGGAUACUGGGGAUUGUGAACCUACAAAAGCGAGUGCCUAGCCAGUUCGUGGGGCAUCAGUUUGGAGUAGGUGGUUCCG -----(.((((((((.(((((.(....)))))))))))..((((((..((((..((((((.((......)).))))))..))))...))))))..)))). ( -29.50) >DroSim_CAF1 2543 95 + 1 -----GUGAGCUAAUACUCCAGGGAUACUGGGGAUUGUGAACCUACAAAAGCGAGUGCCUGGCCAGUUCGUGGGGCAUCAGCUUGGAGUAGGUGGUUCCG -----(.(((((....((((((.....)))))).......((((((..((((..((((((.((......)).))))))..))))...)))))))))))). ( -40.30) >DroEre_CAF1 2763 95 + 1 -----AUGAGGAAACACUCCAAGGAUACUAAGGAUAGUAGACCUACAAAAGCGAGUGCCUGGCCAGUUCGUGGGGCAUCAGCUUGGAGUAGGUGGUUCCG -----....((((.((((((.(((.(((((....)))))..)))....((((..((((((.((......)).))))))..))))))))).)....)))). ( -34.80) >DroYak_CAF1 2548 95 + 1 -----AUGAGAAAAUUCUUCAUGGAUACUAAGGAUUGUGAACCUACAAAAGCGAGUGCCUGGCCAGUUCGUGGGGCAUCAGCUUGGAGUAGGUGGUUCCG -----((((((....)).)))).........(((......((((((..((((..((((((.((......)).))))))..))))...))))))...))). ( -31.00) >DroAna_CAF1 1832 97 + 1 AAAAAAUUGGAAAAUAUUUCAAAAUAAA---UCAACAAAAACCUACAGAAGCGAGUGCCUGGCCAGCUCAUGGGGCAUCAGCUUGGAAUAUGUCGUCCCG ........(((..(((((((........---...........(....)((((..(((((...(((.....))))))))..)))))))))))....))).. ( -20.40) >consensus _____AUGAGAAAAUACUCCAAGGAUACUGGGGAUUGUAAACCUACAAAAGCGAGUGCCUGGCCAGUUCGUGGGGCAUCAGCUUGGAGUAGGUGGUUCCG ...............................(((......((((((..((((..((((((.((......)).))))))..))))...))))))...))). (-23.50 = -24.53 + 1.03)

| Location | 19,375,611 – 19,375,706 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -17.09 |
| Energy contribution | -17.48 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
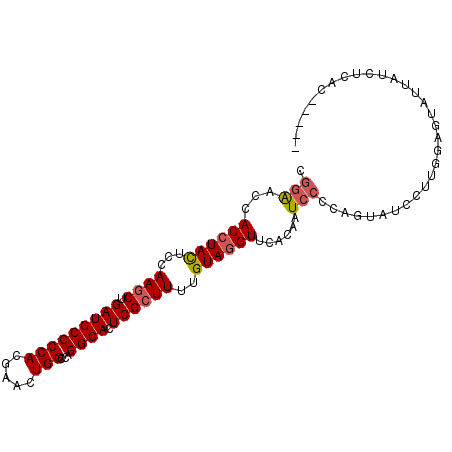
>2R_DroMel_CAF1 19375611 95 - 20766785 CGGAACCACCUACUCCAAGCUGAUGCCCCAUGAACUGGCCAGGCACUCGCUUUUGUAGGUUUACAAUCCCCAAUAUCCUUGGAGUAUUAGCUUAC----- .(((.........)))((((((((((....((((((.((.((((....))))..)).))))))......((((.....)))).))))))))))..----- ( -24.10) >DroSec_CAF1 2492 95 - 1 CGGAACCACCUACUCCAAACUGAUGCCCCACGAACUGGCUAGGCACUCGCUUUUGUAGGUUCACAAUCCCCAGUAUCCUUGGAGUAUUAUCUCAC----- .((.....))((((((((((((.........(((((.((.((((....))))..)).)))))........))))....)))))))).........----- ( -21.43) >DroSim_CAF1 2543 95 - 1 CGGAACCACCUACUCCAAGCUGAUGCCCCACGAACUGGCCAGGCACUCGCUUUUGUAGGUUCACAAUCCCCAGUAUCCCUGGAGUAUUAGCUCAC----- .(((.........))).(((((((((.....(((((.((.((((....))))..)).))))).......((((.....)))).)))))))))...----- ( -26.30) >DroEre_CAF1 2763 95 - 1 CGGAACCACCUACUCCAAGCUGAUGCCCCACGAACUGGCCAGGCACUCGCUUUUGUAGGUCUACUAUCCUUAGUAUCCUUGGAGUGUUUCCUCAU----- .(((...((((((...((((.(((((((((.....)))...)))).))))))..)))))).(((((....))))))))...(((......)))..----- ( -26.60) >DroYak_CAF1 2548 95 - 1 CGGAACCACCUACUCCAAGCUGAUGCCCCACGAACUGGCCAGGCACUCGCUUUUGUAGGUUCACAAUCCUUAGUAUCCAUGAAGAAUUUUCUCAU----- .(((...((((((...((((.(((((((((.....)))...)))).))))))..))))))..((........)).)))((((.((....))))))----- ( -22.10) >DroAna_CAF1 1832 97 - 1 CGGGACGACAUAUUCCAAGCUGAUGCCCCAUGAGCUGGCCAGGCACUCGCUUCUGUAGGUUUUUGUUGA---UUUAUUUUGAAAUAUUUUCCAAUUUUUU .(((((((((.(..((((((.(((((((((.....)))...)))).)))))).....))..).)))))(---(((......))))...))))........ ( -18.70) >consensus CGGAACCACCUACUCCAAGCUGAUGCCCCACGAACUGGCCAGGCACUCGCUUUUGUAGGUUCACAAUCCCCAGUAUCCUUGGAGUAUUAUCUCAC_____ .(((...((((((...((((.(((((((((.....)))...)))).))))))..))))))......)))............................... (-17.09 = -17.48 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:19 2006