
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,373,268 – 19,373,385 |
| Length | 117 |
| Max. P | 0.609558 |

| Location | 19,373,268 – 19,373,385 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.69 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -18.70 |
| Energy contribution | -17.78 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
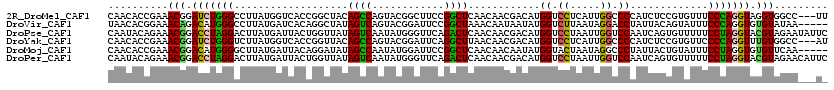
>2R_DroMel_CAF1 19373268 117 + 20766785 CAACACCGAAACGGAUCUGGGCCUUAUGGUCACCGGCUACAGCCAGUACGGCUUCCGGCUCAACAACGACAUGGUCCUCAUUGGCCCCAUCUCCGUGUUUCCCAGGUAGGUGGCC---UU ...((((((..((((.(((((((....))))((.(((....))).)).)))..))))..))((((..((.((((.((.....))..)))).))..)))).........))))...---.. ( -36.10) >DroVir_CAF1 150 115 + 1 UAACACGGAAACAGACAUGGGCCUUAUGAUCACAGGCUAUAGUCAGUACGGAUUCCGGCUAAACAAUAAUAUGGUCUUAAUAGGACCUAUUACAGUAUUUCCCAGGUGUGUAUAA----- ..(((((((((..(((...(((((.........)))))...)))(((.(((...))))))..((..(((((.(((((.....))))))))))..)).)))))...))))......----- ( -27.70) >DroPse_CAF1 147 120 + 1 CAAUACAGAAACGGACCUAGGACUUAUGAUUACUGGUUAUAGUCAAUAUGGGUUCAGACUCAACAACGACAUGGUCCUAAUUGGUCCAAUCAGUGUUUUUCCUAGGUACGUAGAAUAUUC ..........(((.((((((((.....((.((((((((...(((....(((((....))))).....)))..((.((.....)).)))))))))).)).)))))))).)))......... ( -33.80) >DroYak_CAF1 147 117 + 1 CAACACCGAAACGGAUCUGGGUCUUAUGGUCACCGGUUACAGCCAGUACGGAUUCAGGCUUAACAACGACAUGGUCCUCAUUGGCCCCAUCUCCGUGUUCCCCAGGUUUGUGGCC---AU .......(..(((((((((((...(((((.....((.....((((((..((((.((..(........)...))))))..)))))))).....)))))...)))))))))))..).---.. ( -33.70) >DroMoj_CAF1 153 115 + 1 CAACACCGAAACGGACAUGGGGCUUAUGAUUACAGGAUAUAGCCAAUAUGGAUUCCGGCUCAACAACAAUAUGGUACUAAUAGGCCCUAUUACUGUAUUUCCUAGGUGUGUUCAA----- ..((((((((((((..(((((((((((...(((.(((.....((.....))..)))(......).........)))...)))))))))))..))))..)))...)))))......----- ( -30.70) >DroPer_CAF1 147 120 + 1 CAAUACAGAAACGGACCUAGGACUUAUGAUUACUGGUUAUAGUCAAUAUGGGUUCAGACUCAACAACGACAUGGUCCUAAUUGGUCCAAUCAGUGUUUUUCCUAGGUACGUAGAACAUUC ..........(((.((((((((.....((.((((((((...(((....(((((....))))).....)))..((.((.....)).)))))))))).)).)))))))).)))......... ( -33.80) >consensus CAACACCGAAACGGACCUGGGACUUAUGAUCACAGGUUAUAGCCAAUACGGAUUCAGGCUCAACAACGACAUGGUCCUAAUUGGCCCAAUCACUGUAUUUCCCAGGUAUGUAGAA___U_ ..........(((.(((((((...................((((............))))............((.((.....)).)).............))))))).)))......... (-18.70 = -17.78 + -0.91)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:17 2006