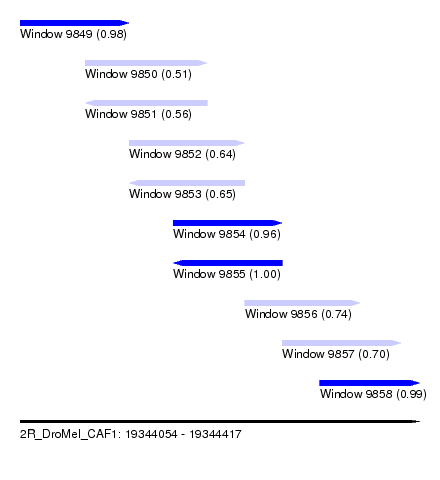
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,344,054 – 19,344,417 |
| Length | 363 |
| Max. P | 0.995707 |

| Location | 19,344,054 – 19,344,153 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 88.28 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.978948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19344054 99 + 20766785 AUCAGCCAGUGUUGCUACUGCUCCAGGCGGCACAAAAGCAUCAACAGCACCCAGUGUUGCAACUGCAGCAGUGGUUAGUGUCUCGUCAGGAGCAACAGU .............(((..((((((.((((((((....((.......))..(((.((((((....)))))).)))...))))..)))).))))))..))) ( -35.60) >DroSec_CAF1 6362 99 + 1 AUCAGCCAGUGUUGCUGGUGCUCCAGGCGGCACAAAAGCAUCAACAGCACCUGGUGUUGCAACUGCAACAGUGGUCACUGUCUCGUCGGGAGCAACAGU ....(((((.....)))))(((((.((((((......))....((((.(((.(.((((((....)))))).))))..))))..)))).)))))...... ( -37.70) >DroSim_CAF1 6318 99 + 1 AUCGGCCAGUGUUGCUGCUGCUCCAGGCGGCACAAAAGCAUCAACAGCACCUGGUGUUGCAACUGCAACAGUGGUCACUGUCUCGUCGGGAGCAACAGU ...(((((.((((((((((((.....)))))).....(((.((.(((...))).)).)))....)))))).)))))((((((((.....)))..))))) ( -39.30) >DroEre_CAF1 6626 99 + 1 AUCGGCCAGUAUUGCUGCUGCUCCAGGCGGCACAAAAGCGUCAACGGCACCUGGUAUUGCAACUGCAACAGUGAUCAGUGUCUCGCCAGGAGCUGCUCC ...(((.......)))((.(((((.((((((......))......(((((.((((.((((....)))).....))))))))).)))).))))).))... ( -33.70) >DroYak_CAF1 6697 99 + 1 AACGGCCAGUGUUGCUGCUGCUUCAGGUGGCACAAAAGCAUCGCCAGCACCUGGUGUUGCAACUGCCACAGUGGUCAGUGUUUCGCCAGGAGCAACAGU ...(((..(((((..(((..(.....)..)))....))))).))).((.(((((((..(((.(((((.....)).))))))..))))))).))...... ( -40.70) >consensus AUCGGCCAGUGUUGCUGCUGCUCCAGGCGGCACAAAAGCAUCAACAGCACCUGGUGUUGCAACUGCAACAGUGGUCAGUGUCUCGUCAGGAGCAACAGU .............((((.((((((.(((((.(((...((.......))..(((.((((((....)))))).)))....))).))))).)))))).)))) (-31.30 = -31.50 + 0.20)

| Location | 19,344,113 – 19,344,224 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -39.67 |
| Consensus MFE | -22.92 |
| Energy contribution | -24.00 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19344113 111 + 20766785 CAACUGCAGCAGUGGUUAGUGUCUCGUCAGGAGCAACAGUGCCUGCCGCAGCCUCAAAAGCAGCCGCUCCACUCAAAGCUACGGCACCACCACCAUCA---------UUAGCUGCGGUUA .(((((((((((((((..(((........(((((.....(((.....)))((.......))....))))).......((....)).....))).))))---------)).))))))))). ( -36.90) >DroSec_CAF1 6421 108 + 1 CAACUGCAACAGUGGUCACUGUCUCGUCGGGAGCAACAGUGCCUGCUGCAGCCUCAAAAGCAGCCGCUCCACUCAAAGCUACGGCACCACCACCAUC------------AGCUGCGGUUA .(((((((.((((((.(((((((((.....)))..))))))...(((((..........)))))....)))))....((....))............------------.).))))))). ( -33.70) >DroSim_CAF1 6377 108 + 1 CAACUGCAACAGUGGUCACUGUCUCGUCGGGAGCAACAGUGCCUGCUGCAGCCUCAAAAGCAGCCGCUCCACUCAAAGCUACGGCACCACCACCAUC------------GGCUGCGGUUA .((((((..(((..(.(((((((((.....)))..)))))).)..)))..)).......(((((((...........((....))...........)------------)))))))))). ( -38.15) >DroEre_CAF1 6685 105 + 1 CAACUGCAACAGUGAUCAGUGUCUCGCCAGGAGCUGCUCCAGUGGCUGGAGCCUCAAAAGCCGCCGCUCCACUCAAAGCCGGGGCU---------------GGUGCUGCAGCUGCGGUUA .(((((((...((...(((..((.......(((..(((((((...))))))))))...((((.(((((........)).)))))))---------------))..)))..))))))))). ( -39.20) >DroYak_CAF1 6756 120 + 1 CAACUGCCACAGUGGUCAGUGUUUCGCCAGGAGCAACAGUGGCGGCUGGUGCCUCAAAAGCAGCCGCUCCACUCAAAGCUACGGCUGCACCAUCAGCCGUUGGUGCUGCAGCUGCGGUGA ..((((((.....)).))))...(((((...(((...(((((((((((.(........).)))))))..))))....))).((((((((.((((((...)))))).)))))))).))))) ( -50.40) >consensus CAACUGCAACAGUGGUCAGUGUCUCGUCAGGAGCAACAGUGCCUGCUGCAGCCUCAAAAGCAGCCGCUCCACUCAAAGCUACGGCACCACCACCAUC____________AGCUGCGGUUA .(((((((.((((((.(((((((((.....)))..)))))).))))))..............((((((........)))...)))...........................))))))). (-22.92 = -24.00 + 1.08)

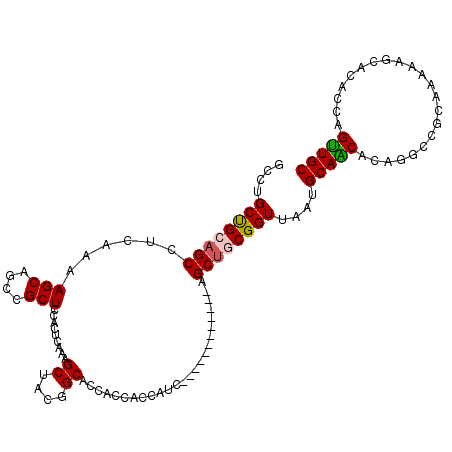
| Location | 19,344,113 – 19,344,224 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -45.88 |
| Consensus MFE | -27.78 |
| Energy contribution | -30.10 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19344113 111 - 20766785 UAACCGCAGCUAA---------UGAUGGUGGUGGUGCCGUAGCUUUGAGUGGAGCGGCUGCUUUUGAGGCUGCGGCAGGCACUGUUGCUCCUGACGAGACACUAACCACUGCUGCAGUUG ((((.(((((...---------.(.((((((((.(((((((((((((((..(.....)..)))..))))))))))))((((....))))..........)))).))))).))))).)))) ( -45.60) >DroSec_CAF1 6421 108 - 1 UAACCGCAGCU------------GAUGGUGGUGGUGCCGUAGCUUUGAGUGGAGCGGCUGCUUUUGAGGCUGCAGCAGGCACUGUUGCUCCCGACGAGACAGUGACCACUGUUGCAGUUG .....((((((------------.(((((......))))).(((((....)))))))))))......(((((((((((.((((((..(((.....)))))))))....))))))))))). ( -45.90) >DroSim_CAF1 6377 108 - 1 UAACCGCAGCC------------GAUGGUGGUGGUGCCGUAGCUUUGAGUGGAGCGGCUGCUUUUGAGGCUGCAGCAGGCACUGUUGCUCCCGACGAGACAGUGACCACUGUUGCAGUUG .....((((((------------.(((((......))))).(((((....)))))))))))......(((((((((((.((((((..(((.....)))))))))....))))))))))). ( -48.60) >DroEre_CAF1 6685 105 - 1 UAACCGCAGCUGCAGCACC---------------AGCCCCGGCUUUGAGUGGAGCGGCGGCUUUUGAGGCUCCAGCCACUGGAGCAGCUCCUGGCGAGACACUGAUCACUGUUGCAGUUG ......(((((((((((..---------------(((((((.((((....))))))).))))..(((.(((((((...))))))).((.....))..........))).))))))))))) ( -41.90) >DroYak_CAF1 6756 120 - 1 UCACCGCAGCUGCAGCACCAACGGCUGAUGGUGCAGCCGUAGCUUUGAGUGGAGCGGCUGCUUUUGAGGCACCAGCCGCCACUGUUGCUCCUGGCGAAACACUGACCACUGUGGCAGUUG (((..((((((((..((((((((((((......))))))).....)).)))..))))))))...)))(((....)))...(((((..(...(((((......)).)))..)..))))).. ( -47.40) >consensus UAACCGCAGCU____________GAUGGUGGUGGUGCCGUAGCUUUGAGUGGAGCGGCUGCUUUUGAGGCUGCAGCAGGCACUGUUGCUCCUGACGAGACACUGACCACUGUUGCAGUUG .....((((((..................((.....))...(((((....)))))))))))......(((((((((((.((((((..(((.....)))))))))....))))))))))). (-27.78 = -30.10 + 2.32)


| Location | 19,344,153 – 19,344,258 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -17.09 |
| Energy contribution | -17.13 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19344153 105 + 20766785 GCCUGCCGCAGCCUCAAAAGCAGCCGCUCCACUCAAAGCUACGGCACCACCACCAUCA---------UUAGCUGCGGUUAAUGCAACACAAGCCGCAAAAAGCACACCAGUUGC ....((((((((......(((....))).........((....)).............---------...))))))))....(((((.......((.....))......))))) ( -25.32) >DroSec_CAF1 6461 102 + 1 GCCUGCUGCAGCCUCAAAAGCAGCCGCUCCACUCAAAGCUACGGCACCACCACCAUC------------AGCUGCGGUUAGUGCAGCACAGGCCGCAAAGAGCACACCAGUUGC ....(((((..........))))).((((........((....))............------------...((((((..(((...)))..))))))..))))........... ( -29.60) >DroSim_CAF1 6417 102 + 1 GCCUGCUGCAGCCUCAAAAGCAGCCGCUCCACUCAAAGCUACGGCACCACCACCAUC------------GGCUGCGGUUAGUGCAACACAGGCCGCCAAGAACACACCAGUUGC (((((.((..((((.((..(((((((...........((....))...........)------------))))))..)))).))..)))))))..................... ( -30.15) >DroEre_CAF1 6725 99 + 1 AGUGGCUGGAGCCUCAAAAGCCGCCGCUCCACUCAAAGCCGGGGCU---------------GGUGCUGCAGCUGCGGUUAAUGCAGCACAGGCCGCAAAAAUCACACCAGCUGC .(..(((((..........((.((((((((..........))))).---------------.((((((((...........)))))))).))).))..........)))))..) ( -39.65) >DroYak_CAF1 6796 114 + 1 GGCGGCUGGUGCCUCAAAAGCAGCCGCUCCACUCAAAGCUACGGCUGCACCAUCAGCCGUUGGUGCUGCAGCUGCGGUGAAUGCAACACAGGUCGCAAAAAUCACACCAGUUGC .((((((((((..(((...(((((.((..(((.((.....(((((((......))))))))))))..)).)))))..))).(((.((....)).))).......)))))))))) ( -48.20) >consensus GCCUGCUGCAGCCUCAAAAGCAGCCGCUCCACUCAAAGCUACGGCACCACCACCAUC____________AGCUGCGGUUAAUGCAACACAGGCCGCAAAAAGCACACCAGUUGC ....((((((((((....))..((((((........)))...))).........................))))))))....(((((......................))))) (-17.09 = -17.13 + 0.04)

| Location | 19,344,153 – 19,344,258 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -43.76 |
| Consensus MFE | -26.10 |
| Energy contribution | -26.58 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19344153 105 - 20766785 GCAACUGGUGUGCUUUUUGCGGCUUGUGUUGCAUUAACCGCAGCUAA---------UGAUGGUGGUGGUGCCGUAGCUUUGAGUGGAGCGGCUGCUUUUGAGGCUGCGGCAGGC (((((.(((.(((.....))))))...)))))....(((((.((((.---------...)))).)))))((((((((((((((..(.....)..)))..))))))))))).... ( -43.40) >DroSec_CAF1 6461 102 - 1 GCAACUGGUGUGCUCUUUGCGGCCUGUGCUGCACUAACCGCAGCU------------GAUGGUGGUGGUGCCGUAGCUUUGAGUGGAGCGGCUGCUUUUGAGGCUGCAGCAGGC ....(((.(((((.(((.((((((..(((.((((((.((((....------------....)))))))))).)))(((((....)))))))))))....))))).))).))).. ( -43.50) >DroSim_CAF1 6417 102 - 1 GCAACUGGUGUGUUCUUGGCGGCCUGUGUUGCACUAACCGCAGCC------------GAUGGUGGUGGUGCCGUAGCUUUGAGUGGAGCGGCUGCUUUUGAGGCUGCAGCAGGC ....(((.((((..((((((((((......((((((.((((....------------....))))))))))....(((((....))))))))))))...)))..)))).))).. ( -40.30) >DroEre_CAF1 6725 99 - 1 GCAGCUGGUGUGAUUUUUGCGGCCUGUGCUGCAUUAACCGCAGCUGCAGCACC---------------AGCCCCGGCUUUGAGUGGAGCGGCGGCUUUUGAGGCUCCAGCCACU (((((((.((.(.((..((((((....))))))..))))))))))))..((..---------------(((((((.((((....))))))).))))..)).(((....)))... ( -40.80) >DroYak_CAF1 6796 114 - 1 GCAACUGGUGUGAUUUUUGCGACCUGUGUUGCAUUCACCGCAGCUGCAGCACCAACGGCUGAUGGUGCAGCCGUAGCUUUGAGUGGAGCGGCUGCUUUUGAGGCACCAGCCGCC ((..(((((((......((((((....))))))((((..((((((((..((((((((((((......))))))).....)).)))..))))))))...)))))))))))..)). ( -50.80) >consensus GCAACUGGUGUGAUUUUUGCGGCCUGUGUUGCAUUAACCGCAGCU____________GAUGGUGGUGGUGCCGUAGCUUUGAGUGGAGCGGCUGCUUUUGAGGCUGCAGCAGGC ......(.((((.....((((((....)))))).....)))).).........................((.(((((((..(...((((....)))))..))))))).)).... (-26.10 = -26.58 + 0.48)

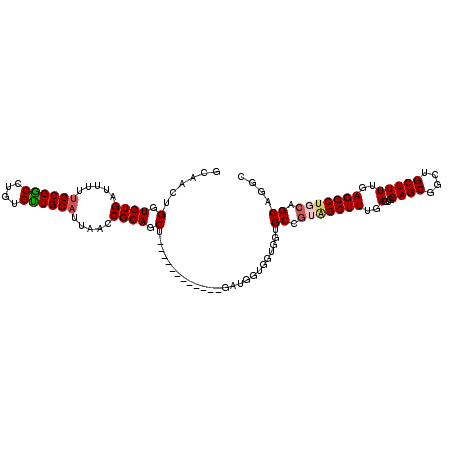
| Location | 19,344,193 – 19,344,292 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.82 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -20.94 |
| Energy contribution | -21.86 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19344193 99 + 20766785 ACGGCACCACCACCAUCA---------UUAGCUGCGGUUAAUGCAACACAAGCCGCAAAAAGCACACCAGUUGCCGCUGCAU------CGGCACUUGUUGCUUCUGCGCCGCCA ..(((...........((---------(((((....)))))))........(.((((..(((((.((.(((.((((......------))))))).))))))).)))).)))). ( -30.40) >DroSec_CAF1 6501 96 + 1 ACGGCACCACCACCAUC------------AGCUGCGGUUAGUGCAGCACAGGCCGCAAAGAGCACACCAGUUGCCGCUCCAU------CGGCACUUGUUGCUGCUGCGCCGCCA ..(((............------------.((((((.....))))))...(((.(((...((((.((.(((.((((......------))))))).))))))..))))))))). ( -36.70) >DroSim_CAF1 6457 102 + 1 ACGGCACCACCACCAUC------------GGCUGCGGUUAGUGCAACACAGGCCGCCAAGAACACACCAGUUGCCGCUCCACUUACCGCGGCACUUGUUGUUGCAGCGCCGCNN .((((.....(......------------)((((((((..(((...)))..)))......((((.((.(((.(((((..........)))))))).)))))))))))))))... ( -35.10) >DroEre_CAF1 6765 93 + 1 GGGGCU---------------GGUGCUGCAGCUGCGGUUAAUGCAGCACAGGCCGCAAAAAUCACACCAGCUGCCGCUCCAU------CGGCACUUGUUGCUACUGCGCCGCCA ((((((---------------((.((.(((((((.((((..(((.((....)).)))..))))....))))))).)).))).------.((((.....)))).....))).)). ( -36.90) >DroYak_CAF1 6836 108 + 1 ACGGCUGCACCAUCAGCCGUUGGUGCUGCAGCUGCGGUGAAUGCAACACAGGUCGCAAAAAUCACACCAGUUGCCGCUCCAU------CGGCAGUUGUUGCUGCUGCACCGCAG .((((.(((.((...((((.(((.((.(((((((..((((.(((.((....)).)))....))))..))))))).)).))).------))))...)).))).))))........ ( -46.50) >consensus ACGGCACCACCACCAUC____________AGCUGCGGUUAAUGCAACACAGGCCGCAAAAAGCACACCAGUUGCCGCUCCAU______CGGCACUUGUUGCUGCUGCGCCGCCA ..(((.........................(.((((.....)))).)...(((.(((...((((.((.((.(((((............))))))).))))))..))))))))). (-20.94 = -21.86 + 0.92)

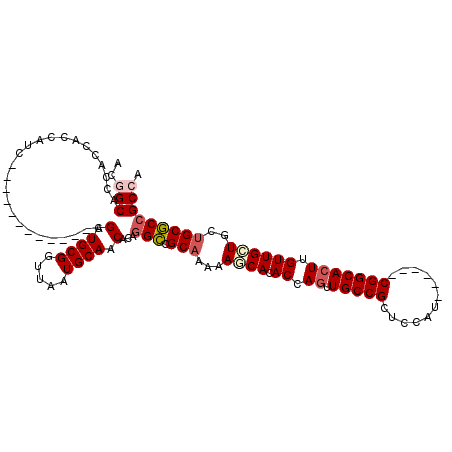

| Location | 19,344,193 – 19,344,292 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.82 |
| Mean single sequence MFE | -45.30 |
| Consensus MFE | -27.08 |
| Energy contribution | -26.28 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19344193 99 - 20766785 UGGCGGCGCAGAAGCAACAAGUGCCG------AUGCAGCGGCAACUGGUGUGCUUUUUGCGGCUUGUGUUGCAUUAACCGCAGCUAA---------UGAUGGUGGUGGUGCCGU ..((((((((((((((.((..(((((------......)))))..))...)))))))((((((....))))))...(((((..(...---------.)...))))).))))))) ( -41.80) >DroSec_CAF1 6501 96 - 1 UGGCGGCGCAGCAGCAACAAGUGCCG------AUGGAGCGGCAACUGGUGUGCUCUUUGCGGCCUGUGCUGCACUAACCGCAGCU------------GAUGGUGGUGGUGCCGU ..(((((((.((((((...((.((((------..((((((.((.....))))))))...)))))).))))))....(((((....------------....))))).))))))) ( -47.70) >DroSim_CAF1 6457 102 - 1 NNGCGGCGCUGCAACAACAAGUGCCGCGGUAAGUGGAGCGGCAACUGGUGUGUUCUUGGCGGCCUGUGUUGCACUAACCGCAGCC------------GAUGGUGGUGGUGCCGU ..(((((((..(.((.....(.((.(((((.((((.((((((..(..(.......)..)..)))...))).)))).))))).)))------------....)).)..))))))) ( -44.50) >DroEre_CAF1 6765 93 - 1 UGGCGGCGCAGUAGCAACAAGUGCCG------AUGGAGCGGCAGCUGGUGUGAUUUUUGCGGCCUGUGCUGCAUUAACCGCAGCUGCAGCACC---------------AGCCCC .((((((((...........))))).------.(((.((.(((((((.((.(.((..((((((....))))))..)))))))))))).)).))---------------)))).. ( -41.80) >DroYak_CAF1 6836 108 - 1 CUGCGGUGCAGCAGCAACAACUGCCG------AUGGAGCGGCAACUGGUGUGAUUUUUGCGACCUGUGUUGCAUUCACCGCAGCUGCAGCACCAACGGCUGAUGGUGCAGCCGU ....(((((.(((((..((..(((((------......)))))..))((((((....((((((....)))))).))).))).))))).))))).(((((((......))))))) ( -50.70) >consensus UGGCGGCGCAGCAGCAACAAGUGCCG______AUGGAGCGGCAACUGGUGUGAUUUUUGCGGCCUGUGUUGCAUUAACCGCAGCU____________GAUGGUGGUGGUGCCGU .((((((((((..(((.((..(((((............)))))..)).))).....)))).)))...(((((.......))))).........................))).. (-27.08 = -26.28 + -0.80)

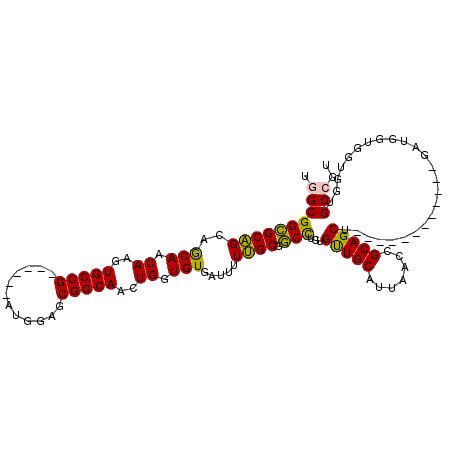
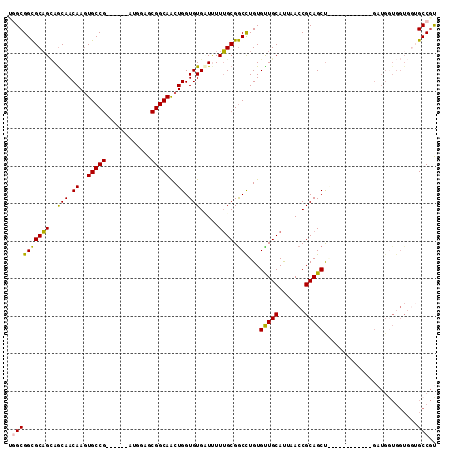
| Location | 19,344,258 – 19,344,363 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.11 |
| Mean single sequence MFE | -31.29 |
| Consensus MFE | -16.39 |
| Energy contribution | -17.59 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19344258 105 + 20766785 CGCUGCAUCGGCACUUGUUGCUUCUGCGCCGCCAGCCAAGUCCA------CAUCAGCAUCGGUGGCGAAGACAAAUACAAACGCUGGCAGCAAGCCAACGGCCACAUUAUC .(((((..((((..((((....(((.(((((((.((........------.....))...))))))).))).....))))..)))))))))..(((...)))......... ( -33.12) >DroSec_CAF1 6563 99 + 1 CGCUCCAUCGGCACUUGUUGCUGCUGCGCCGCCAGCCA------------CAUCAGCAUCGGUGGCGAAGACAAAUACAAACGCUGGCAGCAAGCCAACGGCCGCAUUAUC ........((((..((((...((((.(((((((.((..------------.....))...))))))).)).))...)))).((.((((.....)))).))))))....... ( -32.20) >DroEre_CAF1 6824 105 + 1 CGCUCCAUCGGCACUUGUUGCUACUGCGCCGCCAGCCAAGUCCA------CAUCAGCAUCGGUGGCGAAGACAAAUACCAACGCUGGCAGCAAGCCAGCGGCCGCAUCAAC ........((((..(((((((....))((((((.((........------.....))...))))))...))))).......(((((((.....)))))))))))....... ( -36.22) >DroYak_CAF1 6910 111 + 1 CGCUCCAUCGGCAGUUGUUGCUGCUGCACCGCAGGCCAAGUCCACAUCCACAUCAGCAUCGGUGGCAAAGAAAAAUACCAACGCUGGCAGCAAGCCAACGGCUGCUAUGUC .....(((.((((((((((((((((((......((......))..........((((...((((...........))))...))))))))).)).)))))))))))))).. ( -36.80) >DroAna_CAF1 7487 93 + 1 ------AUCAACAGCAAUC---GCCACGCCUCCUGCAA---CCA------CAUCAGUUGCUGCACCAAAAGUAAACACUGAUUCUGGCUCAAAGCCAACGGCUCCUCUAUC ------.............---(((..((.....))..---...------.((((((((((........))))...))))))..((((.....))))..)))......... ( -18.10) >consensus CGCUCCAUCGGCACUUGUUGCUGCUGCGCCGCCAGCCAAGUCCA______CAUCAGCAUCGGUGGCGAAGACAAAUACAAACGCUGGCAGCAAGCCAACGGCCGCAUUAUC .........(((..((((..(((.(((............................))).)))..)))).............((.((((.....)))).)))))........ (-16.39 = -17.59 + 1.20)


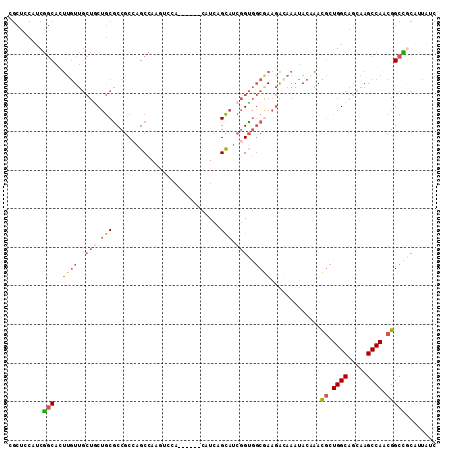
| Location | 19,344,292 – 19,344,400 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.49 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -10.03 |
| Energy contribution | -11.20 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19344292 108 + 20766785 GCCAAGUCCA------CAUCAGCAUCGGUGGCGAAGACAAAUACAAAC---GCUGGCAGCAAGCCAACGGCCACAUUAUCUACUUGCAAGUCGGAUGCU---CCAGCUGCAGCUGCCAGU ((((...((.------..........))))))................---(((((((((..(((...))).............((((.((.((.....---)).))))))))))))))) ( -32.70) >DroSec_CAF1 6597 102 + 1 GCCA------------CAUCAGCAUCGGUGGCGAAGACAAAUACAAAC---GCUGGCAGCAAGCCAACGGCCGCAUUAUCCACUUUCAAGUCGGAUGCU---CCAGCUGCAGCUGCCAGU ((((------------(..........)))))................---(((((((((..(((...))).(((..(((((((....))).))))((.---...))))).))))))))) ( -36.00) >DroEre_CAF1 6858 108 + 1 GCCAAGUCCA------CAUCAGCAUCGGUGGCGAAGACAAAUACCAAC---GCUGGCAGCAAGCCAGCGGCCGCAUCAACUGCACUCAAGUCGGAUGGC---CCAGCUGCAGCAGCAAGU ((((..(((.------((((......)))).....(((.........(---((((((.....)))))))...(((.....)))......))))))))))---...((((...)))).... ( -36.00) >DroWil_CAF1 11612 106 + 1 --CAAGUCAA------CACCAGCAACUGCAACUACCGUAACCAAAAAUGUGUCUGGCACAAAGCCAACUCCUGCUUUGUCCACAUUCAUGCCAGCAAUAAAA------ACAUCAGCAACU --........------.....((...(((.......)))..........((.((((((((((((........)))))))..........)))))))......------......)).... ( -16.60) >DroYak_CAF1 6944 114 + 1 GCCAAGUCCACAUCCACAUCAGCAUCGGUGGCAAAGAAAAAUACCAAC---GCUGGCAGCAAGCCAACGGCUGCUAUGUCCACUUUCAAGUCGGAUGUC---CCAGCUGCAGCAGCAAGU .....................((...((((...........))))...---((((((.....)))..((((((..(((((((((....))).)))))).---.)))))).))).)).... ( -29.00) >DroAna_CAF1 7512 104 + 1 GCAA---CCA------CAUCAGUUGCUGCACCAAAAGUAAACACUGAU---UCUGGCUCAAAGCCAACGGCUCCUCUAUCAACAUUUAAGCCAGAAGCGAAGCCAGCAGCAACCAC---- ....---...------.....((((((((......(((....)))..(---(((((((.((((((...)))).............)).)))))))).........))))))))...---- ( -27.01) >consensus GCCAAGUCCA______CAUCAGCAUCGGUGGCGAAGACAAAUACAAAC___GCUGGCAGCAAGCCAACGGCCGCAUUAUCCACAUUCAAGUCGGAUGCC___CCAGCUGCAGCAGCAAGU .....................((....(((((.....................((((.....)))).((((..................))))............)))))....)).... (-10.03 = -11.20 + 1.17)

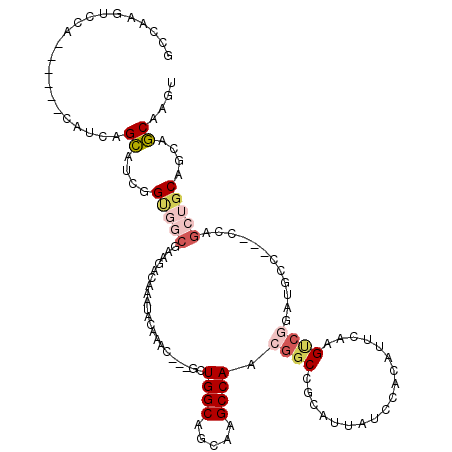
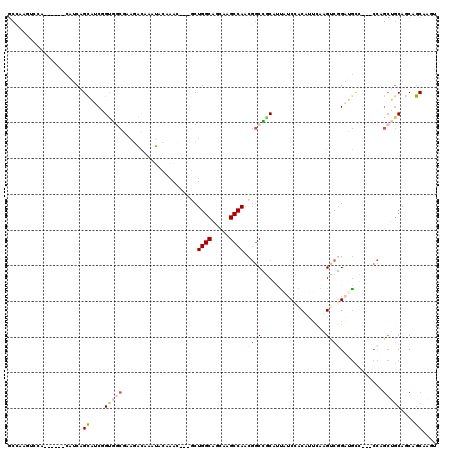
| Location | 19,344,326 – 19,344,417 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 72.61 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -10.60 |
| Energy contribution | -12.28 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.33 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19344326 91 + 20766785 AUACAAAC---GCUGGCAGCAAGCCAACGGCCACAUUAUCUACUUGCAAGUCGGAUGCU---CCAGCUGCAGCUGCCAGUGUUGGGG--UGACGAACGG .(((.(((---(((((((((..(((...))).............((((.((.((.....---)).))))))))))))))))))...)--))........ ( -35.00) >DroSec_CAF1 6625 91 + 1 AUACAAAC---GCUGGCAGCAAGCCAACGGCCGCAUUAUCCACUUUCAAGUCGGAUGCU---CCAGCUGCAGCUGCCAGUGUUGGGG--UGACGAACGG .(((.(((---(((((((((..(((...))).(((..(((((((....))).))))((.---...))))).))))))))))))...)--))........ ( -35.70) >DroEre_CAF1 6892 91 + 1 AUACCAAC---GCUGGCAGCAAGCCAGCGGCCGCAUCAACUGCACUCAAGUCGGAUGGC---CCAGCUGCAGCAGCAAGUGUUGGUG--UGACGAACGG ((((((((---((((((.....))))))(((((..((.(((.......))).)).))))---)..((((...))))....)))))))--)......... ( -35.10) >DroWil_CAF1 11644 93 + 1 CCAAAAAUGUGUCUGGCACAAAGCCAACUCCUGCUUUGUCCACAUUCAUGCCAGCAAUAAAA------ACAUCAGCAACUGUUGCAGGCCCAUCAACAG ......(((.((.(((.(((((((........)))))))))).)).)))(((.((((((...------...........)))))).))).......... ( -21.94) >DroYak_CAF1 6984 91 + 1 AUACCAAC---GCUGGCAGCAAGCCAACGGCUGCUAUGUCCACUUUCAAGUCGGAUGUC---CCAGCUGCAGCAGCAAGUGUUGGUG--CGACGAACGG .(((((((---((((((.....)))..((((((..(((((((((....))).)))))).---.))))))........))))))))))--.......... ( -34.10) >consensus AUACAAAC___GCUGGCAGCAAGCCAACGGCCGCAUUAUCCACUUUCAAGUCGGAUGCC___CCAGCUGCAGCAGCAAGUGUUGGGG__UGACGAACGG ...........((((.((((..((.....)).....((((((((....))).)))))........))))))))......((((.(.......).)))). (-10.60 = -12.28 + 1.68)

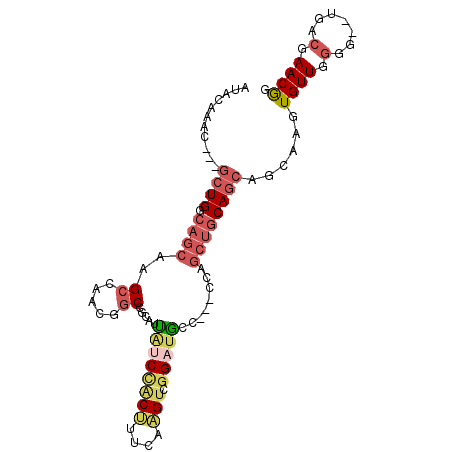
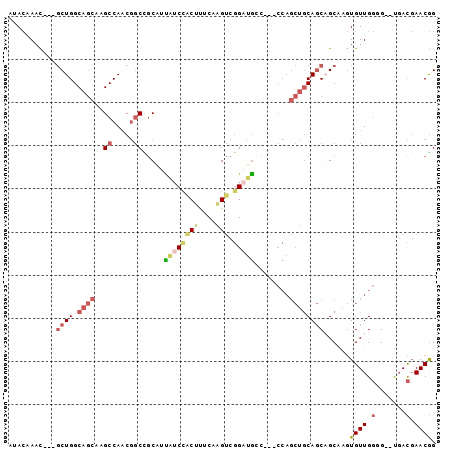
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:08 2006