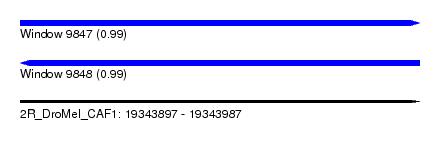
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,343,897 – 19,343,987 |
| Length | 90 |
| Max. P | 0.993709 |

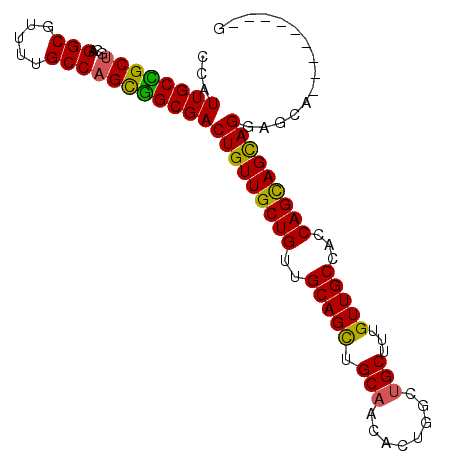
| Location | 19,343,897 – 19,343,987 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 84.83 |
| Mean single sequence MFE | -36.15 |
| Consensus MFE | -28.06 |
| Energy contribution | -29.38 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19343897 90 + 20766785 C---------UGCCACUGCUGCUGGUGGCAACAAAGCAGCCAGUGUUGCAGCUGCAACAGCAACAGUCGCCGCUGGCAAAACGCCUGCAGCAGCAAUGG .---------..(((.(((((((((((((......(((((....))))).((((...))))....))))))((.(((.....))).))))))))).))) ( -42.30) >DroSec_CAF1 6205 90 + 1 C---------AGCUCCUACUACUGGUGGCAACAAAGCACACAAUGUUGCAGCUGCAACAGCAACAGUCGCCACUGGCAAAACGCCUGCAGCGGCAAUGG .---------.((...........(((((......)).)))..((((((....))))))))....(((((..(.(((.....))).)..)))))..... ( -27.50) >DroSim_CAF1 6161 90 + 1 C---------AGCUCCUACUGCUGGUGGCAACAAAGCAGCCAGUGUUGCAGCUGCAACAGCAACAGUCGCCACUGGCAAAACGCCUGCAGCGGCAAUGG .---------.(((....((((.((((((......)).(((((((..((.((((.........)))).)))))))))....)))).)))).)))..... ( -34.20) >DroYak_CAF1 6534 96 + 1 CUGUAACUGCUA---CUGCUGCUGGUGGCAACAAAGCAGCAAGUGUGGCAACUGCAACAGCAACAGUCGCUGCUGGCAAAACGCCUGCUGCAGCAAUGG ((((...(((((---((((((((..((....)).)))))))...))))))..(((....)))))))..(((((.(((.....)))....)))))..... ( -40.60) >consensus C_________AGCUCCUACUGCUGGUGGCAACAAAGCAGCCAGUGUUGCAGCUGCAACAGCAACAGUCGCCACUGGCAAAACGCCUGCAGCAGCAAUGG ................(((((((((((((.((...(((((....))))).((((...))))....)).))))).(((.....)))..)))))))).... (-28.06 = -29.38 + 1.31)

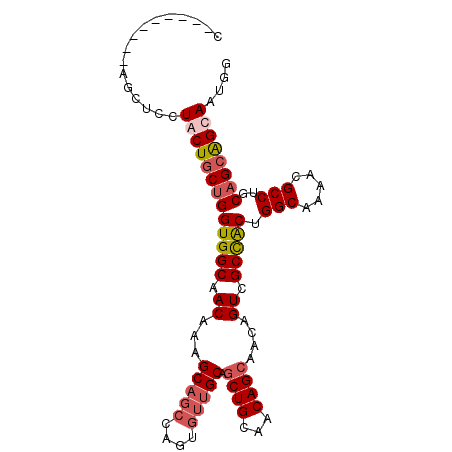
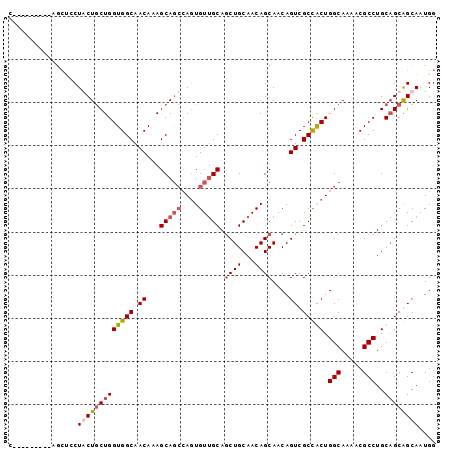
| Location | 19,343,897 – 19,343,987 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 84.83 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -33.44 |
| Energy contribution | -32.62 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19343897 90 - 20766785 CCAUUGCUGCUGCAGGCGUUUUGCCAGCGGCGACUGUUGCUGUUGCAGCUGCAACACUGGCUGCUUUGUUGCCACCAGCAGCAGUGGCA---------G ((((((((((((..((((.......((((((.(.((((((.((....)).)))))).).))))))....))))..))))))))))))..---------. ( -46.20) >DroSec_CAF1 6205 90 - 1 CCAUUGCCGCUGCAGGCGUUUUGCCAGUGGCGACUGUUGCUGUUGCAGCUGCAACAUUGUGUGCUUUGUUGCCACCAGUAGUAGGAGCU---------G ......(((((((.(((.....))).((((((((.(((((....))))).(((........)))...))))))))..))))).))....---------. ( -32.80) >DroSim_CAF1 6161 90 - 1 CCAUUGCCGCUGCAGGCGUUUUGCCAGUGGCGACUGUUGCUGUUGCAGCUGCAACACUGGCUGCUUUGUUGCCACCAGCAGUAGGAGCU---------G ......(((((((.(((.....))).((((((((.((.((((((((....)))))...))).))...))))))))..))))).))....---------. ( -37.30) >DroYak_CAF1 6534 96 - 1 CCAUUGCUGCAGCAGGCGUUUUGCCAGCAGCGACUGUUGCUGUUGCAGUUGCCACACUUGCUGCUUUGUUGCCACCAGCAGCAG---UAGCAGUUACAG .....((((((((.(((.....)))((((((....))))))))))))))..........(((((((((((((.....)))))))---.))))))..... ( -39.20) >consensus CCAUUGCCGCUGCAGGCGUUUUGCCAGCGGCGACUGUUGCUGUUGCAGCUGCAACACUGGCUGCUUUGUUGCCACCAGCAGCAGGAGCA_________G ...((((((((...(((.....)))))))))))(((((((((..(((((.(((........)))...)))))...)))))))))............... (-33.44 = -32.62 + -0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:59 2006