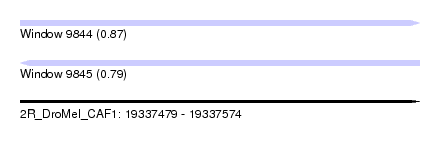
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,337,479 – 19,337,574 |
| Length | 95 |
| Max. P | 0.867392 |

| Location | 19,337,479 – 19,337,574 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.66 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -12.99 |
| Energy contribution | -12.93 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
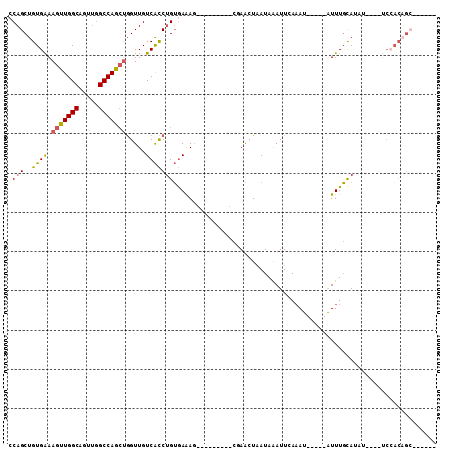
>2R_DroMel_CAF1 19337479 95 + 20766785 CCAGCUGUGAAAGUUGGCAGUUGGCCAGCUGGUUGUCACCUGUGAAAG---------CAAACUAAUAGAUUCAAAU-----AUUUGCAUUUCCCGUCCACAGCUGGAGG (((((((((..(((((((.....)))))))(((....))).(.(((((---------((((...............-----.))))).)))).)...)))))))))... ( -32.49) >DroSec_CAF1 15257 75 + 1 CCAGCUGUGAAAGUUGGCAGUUGGCCAGCUGGUUGUCACCUGUGAAAG---------CUAACUAAUAGAUUCAAAU-----AUUUGCAU-------------------- ((((((.....)))))).(((((((..((.(((....))).))....)---------)))))).............-----........-------------------- ( -18.50) >DroSim_CAF1 15474 85 + 1 CCAGCUGUGAAAGUUGGCAGUUGGCCAGCUGGUUGUCACCUGUGAAAG---------CGAACUAAUAGAUUCAAAU-----AUUUGCAUUU----UCCACAGC------ ...((((((..(((((((.....))))))).(((.(((....))).))---------)..................-----..........----..))))))------ ( -22.70) >DroEre_CAF1 32644 94 + 1 CCAGCUGUGAAAGUUGGCAGUUGGCCAGCUGGUUGUCACCUGUGAAAGUCAACUAAACGAACUAAUAAAUUGAAAU-----AUUUGCAUAU----UCCACAGC------ ...((((((..(((((((.....)))))))((((((((....)))....)))))......................-----..........----..))))))------ ( -23.20) >DroYak_CAF1 15835 94 + 1 CCAGCUGUGAAAGUUGGCAGUUGGCCAGCGGGUUGUCACCUGUGAGAACCAACUAAACGAACUAAUAAAUUGAUAU-----AUUUGCAUAU----UCCACAGC------ ...((((((...((((((.....)))))).((((.(((....))).))))..........................-----..........----..))))))------ ( -27.40) >DroAna_CAF1 15976 86 + 1 ACAGCCGUGGAAAUUGGCUUUCUGCCAAAUAGUUGUCGUCCGGAAAA------------AGGUUAUAAAUUAAAAGAACUUUUUUCUAUAU----CCAACAG------- .......((((..(((((.....))))).............((((((------------(((((............)))))))))))...)----)))....------- ( -19.30) >consensus CCAGCUGUGAAAGUUGGCAGUUGGCCAGCUGGUUGUCACCUGUGAAAG_________CGAACUAAUAAAUUCAAAU_____AUUUGCAUAU____UCCACAGC______ .(((..((((.(((((((.....))))))).....)))))))................................................................... (-12.99 = -12.93 + -0.05)

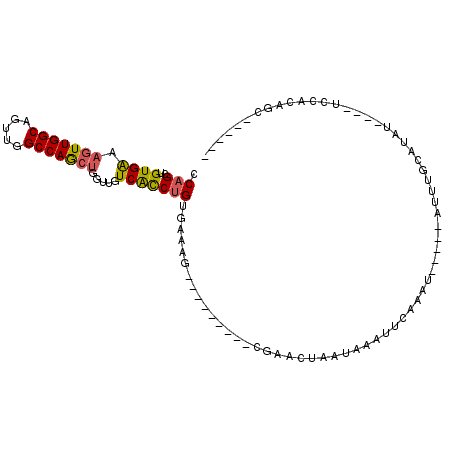
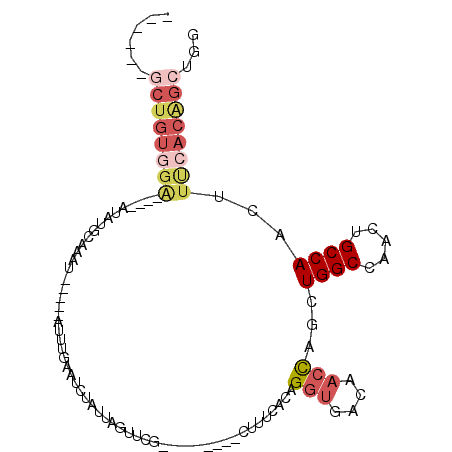
| Location | 19,337,479 – 19,337,574 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 72.66 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -11.50 |
| Energy contribution | -13.03 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19337479 95 - 20766785 CCUCCAGCUGUGGACGGGAAAUGCAAAU-----AUUUGAAUCUAUUAGUUUG---------CUUUCACAGGUGACAACCAGCUGGCCAACUGCCAACUUUCACAGCUGG ...(((((((((((.((((((.((((((-----..............)))))---------)))))...(....)..))((.((((.....)))).))))))))))))) ( -33.54) >DroSec_CAF1 15257 75 - 1 --------------------AUGCAAAU-----AUUUGAAUCUAUUAGUUAG---------CUUUCACAGGUGACAACCAGCUGGCCAACUGCCAACUUUCACAGCUGG --------------------........-----...((((.(((.....)))---------..))))..(....)..(((((((..................))))))) ( -14.97) >DroSim_CAF1 15474 85 - 1 ------GCUGUGGA----AAAUGCAAAU-----AUUUGAAUCUAUUAGUUCG---------CUUUCACAGGUGACAACCAGCUGGCCAACUGCCAACUUUCACAGCUGG ------((((((((----(...((....-----....((((......)))).---------........(((....))).))((((.....))))..)))))))))... ( -23.30) >DroEre_CAF1 32644 94 - 1 ------GCUGUGGA----AUAUGCAAAU-----AUUUCAAUUUAUUAGUUCGUUUAGUUGACUUUCACAGGUGACAACCAGCUGGCCAACUGCCAACUUUCACAGCUGG ------((((((((----(...((....-----...((((((.............))))))........(((....))).))((((.....))))..)))))))))... ( -23.52) >DroYak_CAF1 15835 94 - 1 ------GCUGUGGA----AUAUGCAAAU-----AUAUCAAUUUAUUAGUUCGUUUAGUUGGUUCUCACAGGUGACAACCCGCUGGCCAACUGCCAACUUUCACAGCUGG ------((((((((----(...((.(((-----(........))))((((.(((.(((.((((.(((....))).)))).)))))).))))))....)))))))))... ( -26.50) >DroAna_CAF1 15976 86 - 1 -------CUGUUGG----AUAUAGAAAAAAGUUCUUUUAAUUUAUAACCU------------UUUUCCGGACGACAACUAUUUGGCAGAAAGCCAAUUUCCACGGCUGU -------((((..(----(((..((((((.(((.(........).))).)------------))))).(.....)...))))..))))..((((.........)))).. ( -15.70) >consensus ______GCUGUGGA____AUAUGCAAAU_____AUUUGAAUCUAUUAGUUCG_________CUUUCACAGGUGACAACCAGCUGGCCAACUGCCAACUUUCACAGCUGG ......((((((((.......................................................(((....)))...((((.....))))...))))))))... (-11.50 = -13.03 + 1.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:57 2006