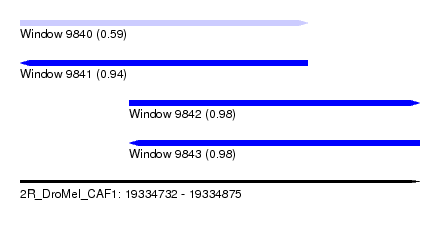
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,334,732 – 19,334,875 |
| Length | 143 |
| Max. P | 0.982471 |

| Location | 19,334,732 – 19,334,835 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -19.58 |
| Consensus MFE | -15.62 |
| Energy contribution | -15.42 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19334732 103 + 20766785 AUAUGCGGGGAAAUUUAACUUUCGCAUAAAAAUGUCAAAAUAGGGAAAAUUAUGAAAUGCCAAAAGGCGGAAAACGCUGUCGGCGGAUUGUUCGCAAAAAAAA .((((((..(.........)..))))))..............(.(((..........((((....((((.....))))...)))).....))).)........ ( -21.26) >DroSec_CAF1 12713 101 + 1 AUAUGCGGGGAAAUUUAACUUUCGCAUAAAAAUGUCAAAAUAGGGAAAACUAUGAAAUGCCAAAAGGCGGAAAACGCUGUCGGCGGAUUGUUCGCAAAA--AA .((((((..(.........)..))))))....(((....((((......))))(((.((((....((((.....))))...)))).....))))))...--.. ( -23.30) >DroSim_CAF1 12890 101 + 1 AUAUGCGGGGAAAUUUAACUUUCGCAUAAAAAUGUCAAAAUAGGGAAAAUUAUGAAAUGCCAAAAGGCGGAAAACGCUGUCGGCGGAUUGUUCGCAAAA--AA .((((((..(.........)..))))))..............(.(((..........((((....((((.....))))...)))).....))).)....--.. ( -21.26) >DroEre_CAF1 12806 94 + 1 A-----GGGA-AAUUUAACUUUCGCAUAAAAAUGUCAAAAUAGGGAAAAUUAUGAAAUGCCAAAAGGCGGAAAACGCUGUCGGCGGAUUGUUCGCAAA---AA .-----(.((-(((((..(((..((((....))))......)))..)))))(..(..((((....((((.....))))...))))..)..))).)...---.. ( -15.70) >DroYak_CAF1 13147 95 + 1 A-----GGGAAAAUUUAACUUUCGCAUAAAAAUGUCAAAAUAGGGAAAAUUAUGAAAUGCCAAAAGGCGGAAAACGCUGUCGGCGGAUUGUUCGCAAA---AA .-----(.((((((((..(((..((((....))))......)))..)))))......((((....((((.....))))...)))).....))).)...---.. ( -16.40) >consensus AUAUGCGGGGAAAUUUAACUUUCGCAUAAAAAUGUCAAAAUAGGGAAAAUUAUGAAAUGCCAAAAGGCGGAAAACGCUGUCGGCGGAUUGUUCGCAAAA__AA ......(.((((((((...((((((((....))))....((((......)))))))).(((....((((.....))))...)))))))).))).)........ (-15.62 = -15.42 + -0.20)

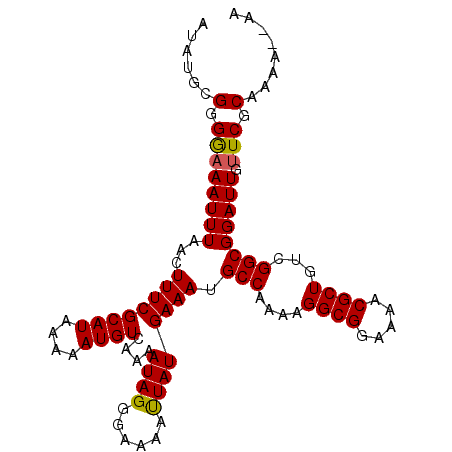
| Location | 19,334,732 – 19,334,835 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -17.05 |
| Consensus MFE | -14.68 |
| Energy contribution | -14.52 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19334732 103 - 20766785 UUUUUUUUGCGAACAAUCCGCCGACAGCGUUUUCCGCCUUUUGGCAUUUCAUAAUUUUCCCUAUUUUGACAUUUUUAUGCGAAAGUUAAAUUUCCCCGCAUAU .......((((........(((((..(((.....)))...)))))......((((((((.(.................).))))))))........))))... ( -18.03) >DroSec_CAF1 12713 101 - 1 UU--UUUUGCGAACAAUCCGCCGACAGCGUUUUCCGCCUUUUGGCAUUUCAUAGUUUUCCCUAUUUUGACAUUUUUAUGCGAAAGUUAAAUUUCCCCGCAUAU ..--...((((........(((((..(((.....)))...)))))......................((.((((....((....)).)))).))..))))... ( -17.80) >DroSim_CAF1 12890 101 - 1 UU--UUUUGCGAACAAUCCGCCGACAGCGUUUUCCGCCUUUUGGCAUUUCAUAAUUUUCCCUAUUUUGACAUUUUUAUGCGAAAGUUAAAUUUCCCCGCAUAU ..--...((((........(((((..(((.....)))...)))))......((((((((.(.................).))))))))........))))... ( -18.03) >DroEre_CAF1 12806 94 - 1 UU---UUUGCGAACAAUCCGCCGACAGCGUUUUCCGCCUUUUGGCAUUUCAUAAUUUUCCCUAUUUUGACAUUUUUAUGCGAAAGUUAAAUU-UCCC-----U ((---((((((........(((((..(((.....)))...)))))...(((.(((.......))).)))........)))))))).......-....-----. ( -15.70) >DroYak_CAF1 13147 95 - 1 UU---UUUGCGAACAAUCCGCCGACAGCGUUUUCCGCCUUUUGGCAUUUCAUAAUUUUCCCUAUUUUGACAUUUUUAUGCGAAAGUUAAAUUUUCCC-----U ((---((((((........(((((..(((.....)))...)))))...(((.(((.......))).)))........))))))))............-----. ( -15.70) >consensus UU__UUUUGCGAACAAUCCGCCGACAGCGUUUUCCGCCUUUUGGCAUUUCAUAAUUUUCCCUAUUUUGACAUUUUUAUGCGAAAGUUAAAUUUCCCCGCAUAU .....((((((........(((((..(((.....)))...)))))...(((.(((.......))).)))........)))))).................... (-14.68 = -14.52 + -0.16)

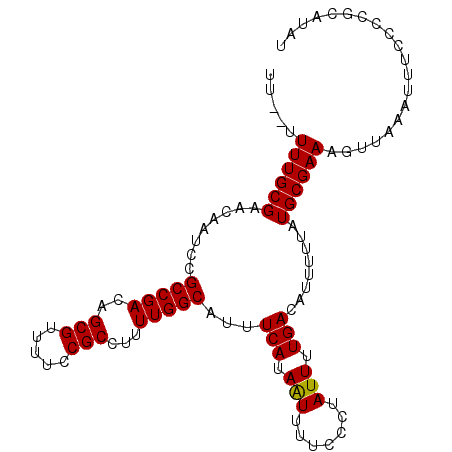
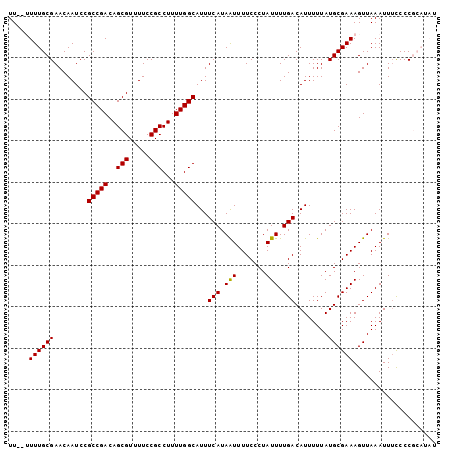
| Location | 19,334,771 – 19,334,875 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 97.41 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -28.22 |
| Energy contribution | -28.04 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.14 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19334771 104 + 20766785 AUAGGGAAAAUUAUGAAAUGCCAAAAGGCGGAAAACGCUGUCGGCGGAUUGUUCGCAAAAAAAAAGGAAGAGAAAGCGAACAGGAAAACUAUAAUGCAGCCCGU ...(((...((((((...((((....((((.....))))...))))..((((((((...................))))))))......))))))....))).. ( -28.51) >DroSim_CAF1 12929 102 + 1 AUAGGGAAAAUUAUGAAAUGCCAAAAGGCGGAAAACGCUGUCGGCGGAUUGUUCGCAAAA--AAAGGAAGAGAAAGUGAACAGGAAAACUAUAAUGCAGCCCAU ...(((...((((((...((((....((((.....))))...))))..((((((((....--.............))))))))......))))))....))).. ( -26.73) >DroEre_CAF1 12839 101 + 1 AUAGGGAAAAUUAUGAAAUGCCAAAAGGCGGAAAACGCUGUCGGCGGAUUGUUCGCAAA---AAAGGAAGAGAAAGCGAACAGGAAAACUAUAAUGCAGCCCAU ...(((...((((((...((((....((((.....))))...))))..((((((((...---.............))))))))......))))))....))).. ( -28.69) >DroYak_CAF1 13181 101 + 1 AUAGGGAAAAUUAUGAAAUGCCAAAAGGCGGAAAACGCUGUCGGCGGAUUGUUCGCAAA---AAAGGAAGAGAAAGCGAACAGGAAAACUAUAAUGCAGCCCAU ...(((...((((((...((((....((((.....))))...))))..((((((((...---.............))))))))......))))))....))).. ( -28.69) >consensus AUAGGGAAAAUUAUGAAAUGCCAAAAGGCGGAAAACGCUGUCGGCGGAUUGUUCGCAAA___AAAGGAAGAGAAAGCGAACAGGAAAACUAUAAUGCAGCCCAU ...(((...((((((...((((....((((.....))))...))))..((((((((...................))))))))......))))))....))).. (-28.22 = -28.04 + -0.19)

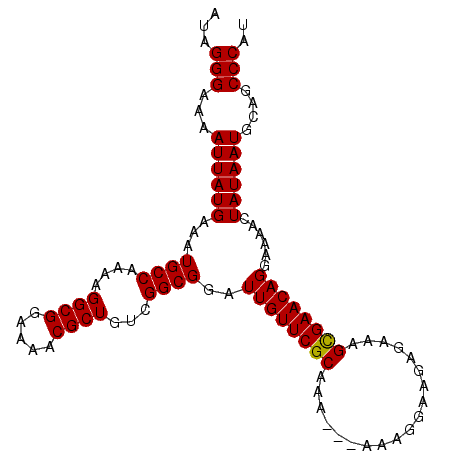
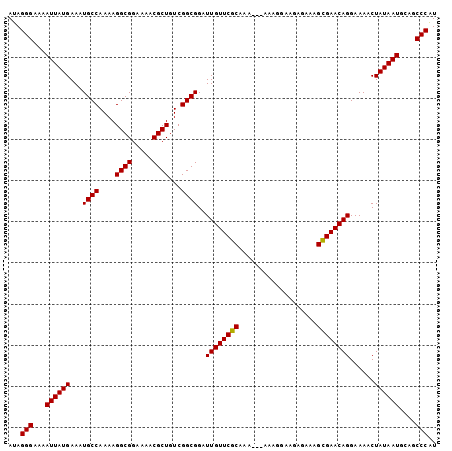
| Location | 19,334,771 – 19,334,875 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 97.41 |
| Mean single sequence MFE | -21.76 |
| Consensus MFE | -21.28 |
| Energy contribution | -21.54 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19334771 104 - 20766785 ACGGGCUGCAUUAUAGUUUUCCUGUUCGCUUUCUCUUCCUUUUUUUUUGCGAACAAUCCGCCGACAGCGUUUUCCGCCUUUUGGCAUUUCAUAAUUUUCCCUAU ..(((....(((((........(((((((...................)))))))....(((((..(((.....)))...))))).....)))))...)))... ( -23.21) >DroSim_CAF1 12929 102 - 1 AUGGGCUGCAUUAUAGUUUUCCUGUUCACUUUCUCUUCCUUU--UUUUGCGAACAAUCCGCCGACAGCGUUUUCCGCCUUUUGGCAUUUCAUAAUUUUCCCUAU ..(((....(((((........(((((.(.............--....).)))))....(((((..(((.....)))...))))).....)))))...)))... ( -17.63) >DroEre_CAF1 12839 101 - 1 AUGGGCUGCAUUAUAGUUUUCCUGUUCGCUUUCUCUUCCUUU---UUUGCGAACAAUCCGCCGACAGCGUUUUCCGCCUUUUGGCAUUUCAUAAUUUUCCCUAU ..(((....(((((........(((((((.............---...)))))))....(((((..(((.....)))...))))).....)))))...)))... ( -23.09) >DroYak_CAF1 13181 101 - 1 AUGGGCUGCAUUAUAGUUUUCCUGUUCGCUUUCUCUUCCUUU---UUUGCGAACAAUCCGCCGACAGCGUUUUCCGCCUUUUGGCAUUUCAUAAUUUUCCCUAU ..(((....(((((........(((((((.............---...)))))))....(((((..(((.....)))...))))).....)))))...)))... ( -23.09) >consensus AUGGGCUGCAUUAUAGUUUUCCUGUUCGCUUUCUCUUCCUUU___UUUGCGAACAAUCCGCCGACAGCGUUUUCCGCCUUUUGGCAUUUCAUAAUUUUCCCUAU ..(((....(((((........(((((((...................)))))))....(((((..(((.....)))...))))).....)))))...)))... (-21.28 = -21.54 + 0.25)


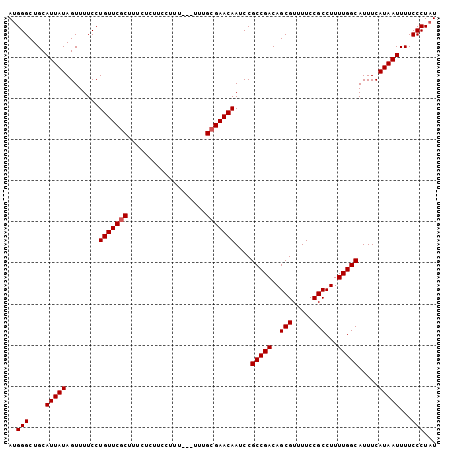
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:55 2006