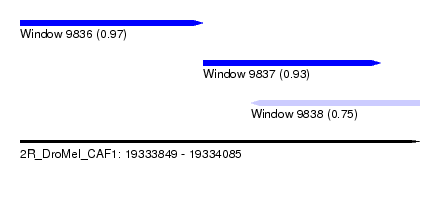
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,333,849 – 19,334,085 |
| Length | 236 |
| Max. P | 0.967275 |

| Location | 19,333,849 – 19,333,957 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -39.75 |
| Consensus MFE | -27.19 |
| Energy contribution | -26.75 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
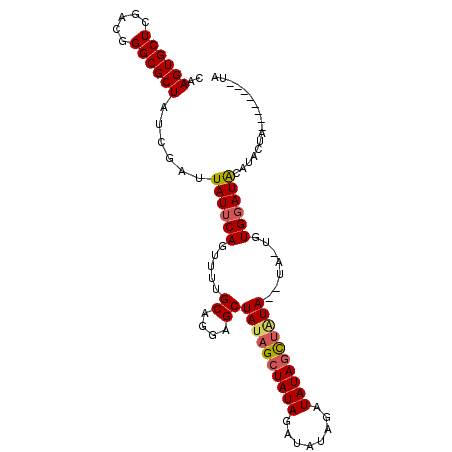
>2R_DroMel_CAF1 19333849 108 + 20766785 --GGCAGCGGGCGGUGCAAUAGAACGUGAAAAUCUGUUUUCCUAUAGCAUUUUCCGGGUGCG----------GGAAAAGCUUGGGAAAGCAGGGGCAACUUCGUUGCAUCGGUACAAAUU --.......(.(((((((((.(((.((.....((((((((((((.(((.(((((((....))----------))))).)))))))))))))))....)))))))))))))).)....... ( -43.10) >DroSec_CAF1 11809 108 + 1 --GGCAGCGGGCGGUGCAAUAGAACGUGAAAAUCUGUUUUCCUCCAGCAUUUUCCGGGUGCG----------GGGAAAGCUUGGGAAAGCAGGGGCAACUUCGUUGCAUCGGUACAAAUU --.......(.(((((((((.(((.((.....(((((((((((..(((.((..(((....))----------)..)).))).)))))))))))....)))))))))))))).)....... ( -41.10) >DroSim_CAF1 11895 108 + 1 --GGCAGCGGGCGGUGCAAUAGAACGUGAAAAUCUGUUUUCCUCUAGCAUUUUCCGGGUGCG----------GGGAAAGCUUGGGAAAGCAGGGGCAACUUCGUUGCAUCGCUACAAAUU --.......(((((((((((.(((.((.....(((((((((((..(((.((..(((....))----------)..)).))).)))))))))))....))))))))))))))))....... ( -46.50) >DroEre_CAF1 11939 117 + 1 GGUGGAGCAGACGAUGCAAUAGAACGUGAAAAUCUGUUUUCCUCCAGCAUUUUCCGGGGGCAGGGGCAG-CUGGAAAAGCUUGGGAA--CAGGGGCAACUUCGUUGCAUCGCCACAAACU .((((......(((((((((.(((.((.....(((((((.((...(((.((((((((..((....))..-))))))))))).)))))--))))....))))))))))))))))))..... ( -43.30) >DroYak_CAF1 12174 116 + 1 --GGCAGCAGGCGAUGCAAUAGAACGUGAAAAUCUGUUUUCCUCUAGCAUUUUCCGGGGGGAGGAGAGGACAGGAGAAGCUGGGGAA--CAGAGGUAACUUCGUUGCAUCAAGACAAAUU --.((.....))((((((((.(((.((.....(((((..(((.(((((.((((((..(............).)))))))))))))))--))))....))))))))))))).......... ( -36.50) >DroAna_CAF1 13069 108 + 1 --GGCUAGGAGCGAUGCAAUAGAACGUGAAAAUCUGUUUUCCGCAGGCGUUUCUAUGGCUUA------AA--CAGAAAAGUAGGGAA--CGGUGACAACUUCGUUGCAUCGAAACAAAUU --.....(...(((((((((.(((..((....(((((.....)))))(((((((...((((.------..--.....)))).)))))--))....))..))))))))))))...)..... ( -28.00) >consensus __GGCAGCGGGCGAUGCAAUAGAACGUGAAAAUCUGUUUUCCUCUAGCAUUUUCCGGGUGCA__________GGAAAAGCUUGGGAA__CAGGGGCAACUUCGUUGCAUCGAUACAAAUU ...........(((((((((.(((.((.....((((..((((.(.(((.((((((.................))))))))).)))))..))))....))))))))))))))......... (-27.19 = -26.75 + -0.44)


| Location | 19,333,957 – 19,334,062 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.85 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -11.04 |
| Energy contribution | -12.84 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19333957 105 + 20766785 UGUGUGCGUGCGCUUGGCCUC-----------AUGCAGCUAUACGAGUAUAGCAUGUAUCCACACUA--UAUAGCUAUAUCUAUAUCUAUAGCUUUAGCUCCUGCAAAACUGAAUAAU .(((((.((((((.((....)-----------).)).((((((....))))))..)))).)))))..--((.(((((((........))))))).))((....))............. ( -27.80) >DroSec_CAF1 11917 96 + 1 UGUGUGCGUGCGCUUGGCCUC-----------AUGCAGCUA--------UAGUAUGUAUCCACA-UA--UAUAGCUAUAUCUAUAUCUAUAGCUAUAGCUCCUGCAAAACUGAAUAAU (((((((((((...((((...-----------.....))))--------..)))))))..))))-..--((((((((((........))))))))))((....))............. ( -25.70) >DroSim_CAF1 12003 96 + 1 UGUGUGCGUGCGCUUGGCCUC-----------AUGCAACUA--------UAGUAUGUAUCCACA-UG--UAUAGCUAUAUCUAUAUCUAUAGCUAUAGCUCCUGCAAAACUGAAUAAU ....((((((((..(((...(-----------((((.....--------..)))))...)))..-))--)))(((((((.((((....)))).)))))))...)))............ ( -23.50) >DroEre_CAF1 12056 100 + 1 UGUGUGCGUGCGCUUGGCCUCUGUCGAUGGUCACGCAGCUA--------U---AUGUAUCCACA-------CAGAUAUAUCUAUAUCUAUAUCUAUAGCUCACGCAAAACUGAAUAAU ....((((((.((.(((((((....)).))))).))(((((--------(---(.((((.....-------.((((((....)))))))))).)))))))))))))............ ( -29.60) >DroYak_CAF1 12290 106 + 1 UGUGUGCGUGCGCUUGGCCUCCGUCGAUGGUCAUCCACACA--------U---AUACAUACAUA-UAUCUACGAGUAUAUCUAUAUCUAUAGCUAUAGCUCCCGCAAAACUGAAUAAU ((((((.(((.(..(((((((....)).)))))..).))).--------)---)))))......-.......(((((((.((((....)))).))).))))................. ( -22.00) >consensus UGUGUGCGUGCGCUUGGCCUC___________AUGCAGCUA________UAG_AUGUAUCCACA_UA__UAUAGCUAUAUCUAUAUCUAUAGCUAUAGCUCCUGCAAAACUGAAUAAU ...((...((((...(((..............(((((((((((....)))))).))))).............(((((((........)))))))...)))..))))..))........ (-11.04 = -12.84 + 1.80)

| Location | 19,333,985 – 19,334,085 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 85.66 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -18.88 |
| Energy contribution | -19.38 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19333985 100 - 20766785 CAAGUGCUCGACGGGCGCUAUCGAUUAUUCAGUUUUGCAGGAGCUAAAGCUAUAGAUAUAGAUAUAGCUAUA--UAGUGUGGAUACAUGCUAUACUCGUAUA ..((((((.....))))))..(((......(((((.....)))))..(((((((........))))))).((--(((((((....))))))))).))).... ( -26.70) >DroSec_CAF1 11945 91 - 1 CAAGUGCUCGACGGGCGCUAUUGAUUAUUCAGUUUUGCAGGAGCUAUAGCUAUAGAUAUAGAUAUAGCUAUA--UA-UGUGGAUACAUACUA--------UA ..((((((.....))))))(((((....)))))((..((..(((((((.((((....)))).)))))))...--..-))..)).........--------.. ( -24.20) >DroSim_CAF1 12031 91 - 1 CAAGUGCUCGACGGGCGCUAUCGAUUAUUCAGUUUUGCAGGAGCUAUAGCUAUAGAUAUAGAUAUAGCUAUA--CA-UGUGGAUACAUACUA--------UA ..((((((.....))))))............((((..(((.(((((((.((((....)))).)))))))...--).-))..)).))......--------.. ( -23.30) >DroYak_CAF1 12329 90 - 1 CAAGUGCUCGACGGGCGCUAUCGAUUAUUCAGUUUUGCGGGAGCUAUAGCUAUAGAUAUAGAUAUACUCGUAGAUA-UAUGUAUGUAU---A--------UG ..((((((.....))))))(((((....))((((..((....))...))))...)))....((((((.((((....-))))...))))---)--------). ( -20.70) >consensus CAAGUGCUCGACGGGCGCUAUCGAUUAUUCAGUUUUGCAGGAGCUAUAGCUAUAGAUAUAGAUAUAGCUAUA__UA_UGUGGAUACAUACUA________UA ..((((((.....))))))......((((((.....((....))((((((((((........)))))))))).......))))))................. (-18.88 = -19.38 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:50 2006