
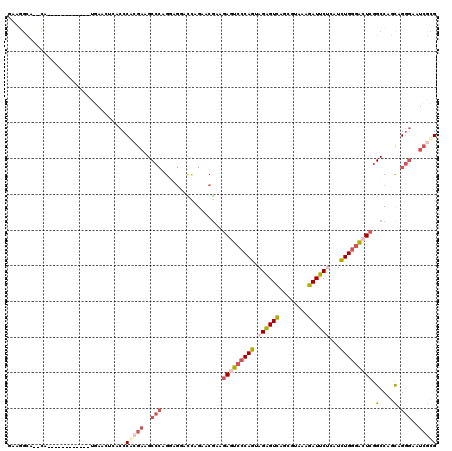
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,332,242 – 19,332,405 |
| Length | 163 |
| Max. P | 0.930601 |

| Location | 19,332,242 – 19,332,348 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.46 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -22.76 |
| Energy contribution | -24.27 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
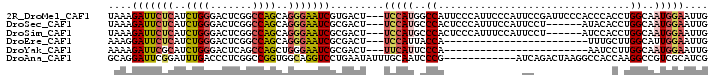
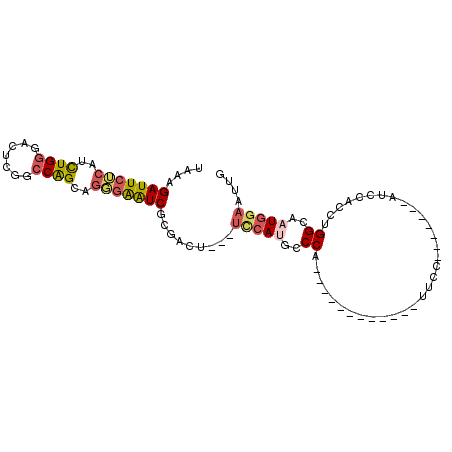
>2R_DroMel_CAF1 19332242 106 + 20766785 GAAGGCA--CA------------UGAACUCACCCACGAAGCCCAGAAGGACCAGAACGAAGAGUCCCAGUAGGGUCAGCGUAAAGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGUG .......--..------------..........(((((..(((.(..((.((........(((((((((.((((((........))))))...)))))))))))))..).)))..))))) ( -36.30) >DroSec_CAF1 10233 106 + 1 GAAGGCA--CA------------CGAACUCACCCACGAAGCCCAGGAGGACCAGAACGAAGAGUCCCAGUAGGGUCAGCGUAAAGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGCG ....((.--..------------.(....).(((.....(((..((....))........(((((((((.((((((........))))))...)))))))))))).....)))....)). ( -33.50) >DroSim_CAF1 10285 106 + 1 GAAGGCA--CA------------UGAACUCACCCAAGAAGCCCAGGAGGAUUAGGACGAAGAGUCCCAGUAGGGUCAGCGUAAAGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGCG ....((.--..------------((....))(((.......((....))....((.....(((((((((.((((((........))))))...)))))))))..))....)))....)). ( -32.20) >DroEre_CAF1 10330 106 + 1 GAAGGCG--CU------------UGAACUCACCCGCGAGGCCCAGGAGAGUCAGGACGAAGAGUCCCAGCAGAGUCAGCGAAAGGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGCG ...((((--((------------(((.(((..((....))(....).)))))))).)...(((((((((.((((((..(....)))))))...))))))))).))).((........)). ( -41.70) >DroYak_CAF1 10551 107 + 1 AAAUGCU--AUC-----------UUAACUCACCCGCGAAGCCCAGAAGAACCAGAACGAAGACUCCCAGCAGAGUCAGCGAAAAGAUUCGCAUCUGGGACUCAGCCAGCUGGGAAUCGCG .......--...-----------..........(((((..(((((.((..(((((.....(((((......))))).(((((....))))).)))))..)).......)))))..))))) ( -36.90) >DroAna_CAF1 11400 120 + 1 UAAAAUAUUUUCAUUUUCAUCAAAAAACUCACCCAAACAGCCGAGGAACAUAAGAAAACCAAUGCCCAGGAGAGUUAACAGCAGGAUUCGGAUUUGACCCUCGGCCGGUGGCAGGUCCUG ............................(((((......(((((((............((........)).(((((........))))).........))))))).)))))......... ( -21.60) >consensus GAAGGCA__CA____________UGAACUCACCCACGAAGCCCAGGAGGACCAGAACGAAGAGUCCCAGUAGAGUCAGCGUAAAGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGCG .................................(((((..(((.................(((((((((.((((((........))))))...))))))))).(....).)))..))))) (-22.76 = -24.27 + 1.50)


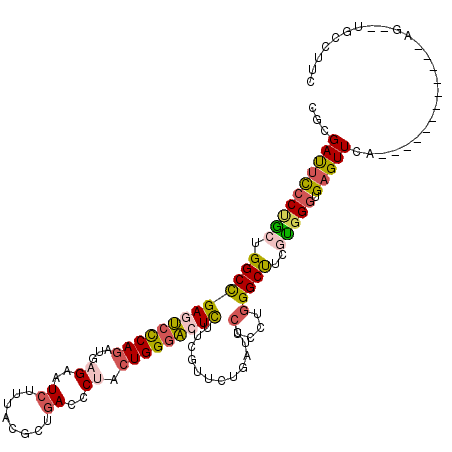
| Location | 19,332,242 – 19,332,348 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.46 |
| Mean single sequence MFE | -36.79 |
| Consensus MFE | -23.19 |
| Energy contribution | -23.78 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19332242 106 - 20766785 CACGAUUCCCUGCUGGCCGAGUCCCAGAUGAGAAUCUUUACGCUGACCCUACUGGGACUCUUCGUUCUGGUCCUUCUGGGCUUCGUGGGUGAGUUCA------------UG--UGCCUUC (((((..(((.(..(((((((((((((...((..((........))..)).)))))))))........)).))..).)))..)))))((((......------------..--))))... ( -35.30) >DroSec_CAF1 10233 106 - 1 CGCGAUUCCCUGCUGGCCGAGUCCCAGAUGAGAAUCUUUACGCUGACCCUACUGGGACUCUUCGUUCUGGUCCUCCUGGGCUUCGUGGGUGAGUUCG------------UG--UGCCUUC ((((((((((..(.(((((((((((((...((..((........))..)).)))))))))........((....))..))))..)..)).))).)))------------))--....... ( -39.30) >DroSim_CAF1 10285 106 - 1 CGCGAUUCCCUGCUGGCCGAGUCCCAGAUGAGAAUCUUUACGCUGACCCUACUGGGACUCUUCGUCCUAAUCCUCCUGGGCUUCUUGGGUGAGUUCA------------UG--UGCCUUC .(((.......(((.((((((((((((...((..((........))..)).)))))))))...((((..........))))......))).)))...------------..--))).... ( -33.20) >DroEre_CAF1 10330 106 - 1 CGCGAUUCCCUGCUGGCCGAGUCCCAGAUGAGAAUCCUUUCGCUGACUCUGCUGGGACUCUUCGUCCUGACUCUCCUGGGCCUCGCGGGUGAGUUCA------------AG--CGCCUUC (((((((((((((.(((((((((((((...(((.((........)).))).)))))))))...((....)).......))))..))))).)))))..------------.)--))..... ( -42.10) >DroYak_CAF1 10551 107 - 1 CGCGAUUCCCAGCUGGCUGAGUCCCAGAUGCGAAUCUUUUCGCUGACUCUGCUGGGAGUCUUCGUUCUGGUUCUUCUGGGCUUCGCGGGUGAGUUAA-----------GAU--AGCAUUU (((((..(((((......(((..(((((.(((((....))))).((((((....)))))).....)))))..))))))))..))))).....((((.-----------..)--))).... ( -39.70) >DroAna_CAF1 11400 120 - 1 CAGGACCUGCCACCGGCCGAGGGUCAAAUCCGAAUCCUGCUGUUAACUCUCCUGGGCAUUGGUUUUCUUAUGUUCCUCGGCUGUUUGGGUGAGUUUUUUGAUGAAAAUGAAAAUAUUUUA ...(((..(((..(((((((((..((.....(((((.((((.............))))..))))).....))..)))))))))....)))..)))((((.((....)).))))....... ( -31.12) >consensus CGCGAUUCCCUGCUGGCCGAGUCCCAGAUGAGAAUCUUUACGCUGACCCUACUGGGACUCUUCGUUCUGAUCCUCCUGGGCUUCGUGGGUGAGUUCA____________AG__UGCCUUC ...((((((((((.(((((((((((((...((..((........))..)).)))))))))............(....)))))..))))).)))))......................... (-23.19 = -23.78 + 0.59)

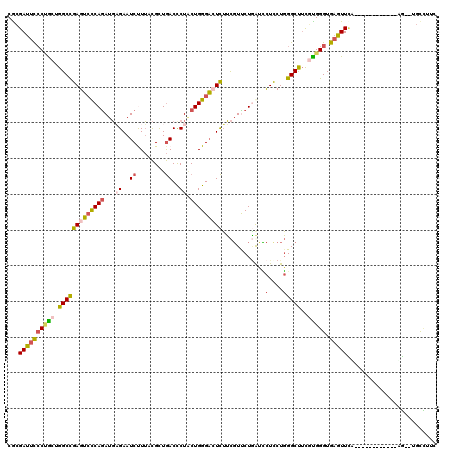
| Location | 19,332,268 – 19,332,385 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.68 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -18.88 |
| Energy contribution | -19.88 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19332268 117 + 20766785 CCCAGAAGGACCAGAACGAAGAGUCCCAGUAGGGUCAGCGUAAAGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGUGACU---UCCAUGGCCAUUCCCAUUCCCAUUCCGAUUCCCA .......((.((........(((((((((.((((((........))))))...)))))))))))))....((((((((.((..---...((((......))))......)))))))))). ( -41.30) >DroSec_CAF1 10259 111 + 1 CCCAGGAGGACCAGAACGAAGAGUCCCAGUAGGGUCAGCGUAAAGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGCGACU---UCCAUGCCCACUCCCAUUUCCAUUCCU------A ...(((((((..........(((((((((.((((((........))))))...)))))))))((...(((.((((.......)---))).))))).........)))..))))------. ( -36.70) >DroSim_CAF1 10311 111 + 1 CCCAGGAGGAUUAGGACGAAGAGUCCCAGUAGGGUCAGCGUAAAGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGCGACU---UCCAUGCCCACUCCCAUUUCCAUUCCU------A ...(((((((...(((....(((((((((.((((((........))))))...)))))))))((...(((.((((.......)---))).)))))..)))....)))..))))------. ( -39.60) >DroEre_CAF1 10356 93 + 1 CCCAGGAGAGUCAGGACGAAGAGUCCCAGCAGAGUCAGCGAAAGGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGCGACU---UCCAUUACCA------------------------ ....(((.((((.((.....(((((((((.((((((..(....)))))))...)))))))))..)).((........))))))---))).......------------------------ ( -35.90) >DroYak_CAF1 10578 93 + 1 CCCAGAAGAACCAGAACGAAGACUCCCAGCAGAGUCAGCGAAAAGAUUCGCAUCUGGGACUCAGCCAGCUGGGAAUCGCGACU---UUCAUUCCCA------------------------ ((((((..............(((((......))))).(((((....))))).))))))............((((((.......---...)))))).------------------------ ( -28.90) >DroAna_CAF1 11440 108 + 1 CCGAGGAACAUAAGAAAACCAAUGCCCAGGAGAGUUAACAGCAGGAUUCGGAUUUGACCCUCGGCCGGUGGCAGGUCCUGAAUAUUUGCAAUCCCG------------AUCAGACUAAGG .((.(((...........((........))..........((((((((((((((((.((.((....)).))))))))).)))).)))))..)))))------------............ ( -24.80) >consensus CCCAGGAGGACCAGAACGAAGAGUCCCAGUAGAGUCAGCGUAAAGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGCGACU___UCCAUGCCCA____________UUCC_______A .......(((..((..(((.(((((((((.((((((........))))))...)))))))))..((....))...)))...))...)))............................... (-18.88 = -19.88 + 1.00)

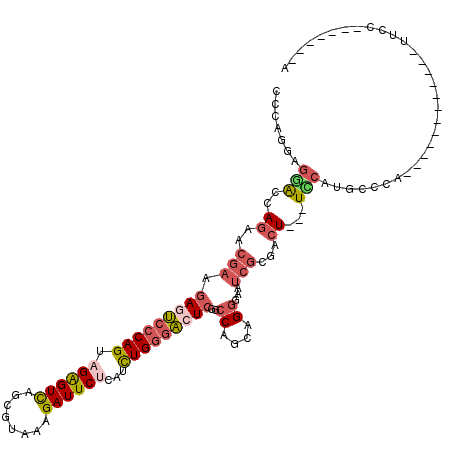

| Location | 19,332,308 – 19,332,405 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 65.16 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -11.52 |
| Energy contribution | -12.13 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19332308 97 + 20766785 UAAAGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGUGACU---UCCAUGGCCAUUCCCAUUCCCAUUCCGAUUCCCACCCACCUGGCAAUGGAAUUG ....((.((((....)))).)).(((((..((((((((.((..---...((((......))))......))))))))))......))))).......... ( -28.10) >DroSec_CAF1 10299 91 + 1 UAAAGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGCGACU---UCCAUGCCCACUCCCAUUUCCAUUCCU------AUACACCUGGCAAUGGAAUUG ....(((((((..((((.......))))..)))))))......---..............((.(((((((((.------........)).))))))).)) ( -22.90) >DroSim_CAF1 10351 91 + 1 UAAAGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGCGACU---UCCAUGCCCACUCCCAUUUCCAUUCCU------AUCCACCUGGCAAUGGAAUUG ....(((((((..((((.......))))..)))))))......---..............((.(((((((((.------........)).))))))).)) ( -22.90) >DroEre_CAF1 10396 73 + 1 AAAGGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGCGACU---UCCAUUACCA------------------------UUUGCUUGGCAUUGGAAUUG ....(((((((..((((.......))))..))))))).....(---((((...(((------------------------......)))...)))))... ( -21.30) >DroYak_CAF1 10618 73 + 1 AAAAGAUUCGCAUCUGGGACUCAGCCAGCUGGGAAUCGCGACU---UUCAUUCCCA------------------------AAUCCUUGGCAAUGGAAUUG ....(((((.((.((((.......)))).)).))))).....(---((((((.(((------------------------(....)))).)))))))... ( -22.50) >DroAna_CAF1 11480 88 + 1 GCAGGAUUCGGAUUUGACCCUCGGCCGGUGGCAGGUCCUGAAUAUUUGCAAUCCCG------------AUCAGACUAAGGCCACCAAGGCCGUCGCAUCG ((((((((((((((((.((.((....)).))))))))).)))).))))).....((------------((..(((...((((.....)))))))..)))) ( -30.30) >consensus UAAAGAUUCUCAUCUGGGACUCGGCCAGCAGGGAAUCGCGACU___UCCAUGCCCA____________UUCC_______AUCCACCUGGCAAUGGAAUUG ....(((((((..((((.......))))..))))))).........(((((..((................................))..))))).... (-11.52 = -12.13 + 0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:45 2006