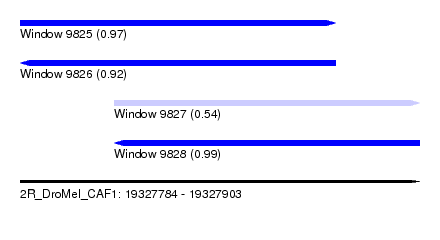
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,327,784 – 19,327,903 |
| Length | 119 |
| Max. P | 0.991107 |

| Location | 19,327,784 – 19,327,878 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 88.51 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -27.24 |
| Energy contribution | -27.92 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19327784 94 + 20766785 AUAAACAUUUAUAGCAAGCGAGCGUGGGUGGAAAAGCUGCGGCGGCGGAGCUUUCCACGCUCCCGAAGCGAAAAGCGUUCGCUCUCGCUUUUCA .............((..(.(((((((((.....(((((.((....)).)))))))))))))).)...))((((((((........)))))))). ( -36.60) >DroSec_CAF1 5723 91 + 1 AUAAGCAUUUAUAGCAAGCGAGCGUGGGUGGAAAAGCUGCGGCGGCGGAGCUUUCCACGCUCCCGAAGCGAAAAGCGCUCGCUCUC---UUCCA ....((.......)).(((((((((..(((((((.(((.((....)).))))))))))(((.....))).....)))))))))...---..... ( -36.00) >DroSim_CAF1 5754 94 + 1 AUAAACAUUUAUAGCAAGCGAGCGUGGGUGGGAAAGCUGCGGCGGCGGAGCUUUCCACGCUCCCGAAGCGAAAAGCGCUCGCUCUCGCUUUCCA ...............(((((((.(((((((((((((((.((....)).)))))))).((((.....)))).....))))))).))))))).... ( -41.60) >DroEre_CAF1 5808 94 + 1 AUAAACAUUUAUGGCAAGCGAGCGUGGGUGGAAAAGCUUCCCGCGCGGAGCUUUCCACGCUCCCGCAAAGAAAAGCGUUCGCUCUCGCUUUCCA ...........(((.(((((((.(((((((((((.(((((......)))))))))))).....(((........))).)))).))))))).))) ( -36.90) >DroYak_CAF1 5771 90 + 1 AUAAACUUUUAUGGCAAGCGAGCGUGGGUGGAAAAGCUGCUGCCGCUGAGCUUUCCACGCUCCCGAAAAGAAAAGCGUCUGCUCUCGCUU---- .....(((((.(((.....(((((((((.((...(((.......)))...)).)))))))))))))))))..(((((........)))))---- ( -29.40) >consensus AUAAACAUUUAUAGCAAGCGAGCGUGGGUGGAAAAGCUGCGGCGGCGGAGCUUUCCACGCUCCCGAAGCGAAAAGCGUUCGCUCUCGCUUUCCA ............(((.((((((((((.(((((((.(((.((....)).)))))))))).)...(.....)....)))))))))...)))..... (-27.24 = -27.92 + 0.68)

| Location | 19,327,784 – 19,327,878 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 88.51 |
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -26.58 |
| Energy contribution | -27.86 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19327784 94 - 20766785 UGAAAAGCGAGAGCGAACGCUUUUCGCUUCGGGAGCGUGGAAAGCUCCGCCGCCGCAGCUUUUCCACCCACGCUCGCUUGCUAUAAAUGUUUAU .((((((((........))))))))((...(.((((((((((((((.((....)).)))))).....)))))))).)..))............. ( -34.40) >DroSec_CAF1 5723 91 - 1 UGGAA---GAGAGCGAGCGCUUUUCGCUUCGGGAGCGUGGAAAGCUCCGCCGCCGCAGCUUUUCCACCCACGCUCGCUUGCUAUAAAUGCUUAU ....(---(..(((((((((((((......))))))((((((((((.((....)).))).)))))))....)))))))..))............ ( -33.70) >DroSim_CAF1 5754 94 - 1 UGGAAAGCGAGAGCGAGCGCUUUUCGCUUCGGGAGCGUGGAAAGCUCCGCCGCCGCAGCUUUCCCACCCACGCUCGCUUGCUAUAAAUGUUUAU ....(((((..(((((((((.....)).....((((((((((((((.((....)).)))))))).....))))))))))))).....))))).. ( -34.90) >DroEre_CAF1 5808 94 - 1 UGGAAAGCGAGAGCGAACGCUUUUCUUUGCGGGAGCGUGGAAAGCUCCGCGCGGGAAGCUUUUCCACCCACGCUCGCUUGCCAUAAAUGUUUAU (((.(((((((.(((((........)))))(((.(((((((....)))))))((((.....)))).)))...))))))).)))........... ( -38.50) >DroYak_CAF1 5771 90 - 1 ----AAGCGAGAGCAGACGCUUUUCUUUUCGGGAGCGUGGAAAGCUCAGCGGCAGCAGCUUUUCCACCCACGCUCGCUUGCCAUAAAAGUUUAU ----((((((((((....))))))........((((((((((((((..((....)))))))).....))))))))))))............... ( -29.70) >consensus UGGAAAGCGAGAGCGAACGCUUUUCGCUUCGGGAGCGUGGAAAGCUCCGCCGCCGCAGCUUUUCCACCCACGCUCGCUUGCUAUAAAUGUUUAU .((((((((........)))))))).....(.((((((((((((((..((....)))))))).....)))))))).)................. (-26.58 = -27.86 + 1.28)

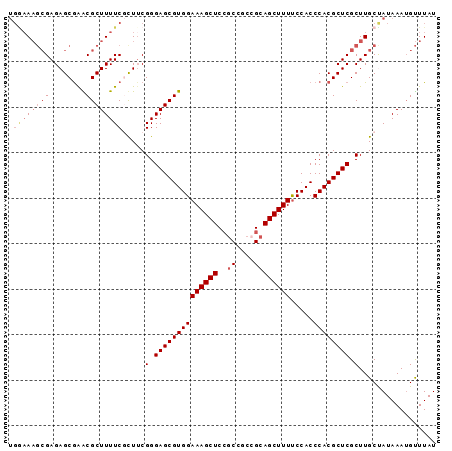
| Location | 19,327,812 – 19,327,903 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 86.02 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -17.12 |
| Energy contribution | -20.32 |
| Covariance contribution | 3.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19327812 91 + 20766785 UGGAAAAGCUGCGGCGGCGGAGCUUUCCACGCUCCCGAAGCGAAAAGCGUUCGCUCUCGCUUUUCACUUUUCACUUUUCUCGCCUCCGCCU----- ..........((((.((((((((.......))))).((((.((((((((........)))))))).))))...........))).))))..----- ( -35.30) >DroSec_CAF1 5751 88 + 1 UGGAAAAGCUGCGGCGGCGGAGCUUUCCACGCUCCCGAAGCGAAAAGCGCUCGCUCUC---UUCCACUUUUCACUUUGCUCGCCUCCGCCU----- ..........((((.((((((((.......)))))(((((.((((((.(.........---...).)))))).)))))...))).))))..----- ( -30.20) >DroSim_CAF1 5782 91 + 1 UGGGAAAGCUGCGGCGGCGGAGCUUUCCACGCUCCCGAAGCGAAAAGCGCUCGCUCUCGCUUUCCACUUUUCACUUUUCUCGCCUCCGCCU----- ..........((((.((((((((.......))))).(((((((..(((....))).)))))))..................))).))))..----- ( -32.30) >DroEre_CAF1 5836 96 + 1 UGGAAAAGCUUCCCGCGCGGAGCUUUCCACGCUCCCGCAAAGAAAAGCGUUCGCUCUCGCUUUCCACUUUUCACUUUUCUCGCCUCCGCCUCCGCC .(((((((......(((.(((((.......))))))))...(.((((((........)))))).)........)))))))................ ( -25.80) >DroYak_CAF1 5799 84 + 1 UGGAAAAGCUGCUGCCGCUGAGCUUUCCACGCUCCCGAAAAGAAAAGCGUCUGCUCUCGCUU-------UUCACUUUUCUCGCCUCCGCCU----- .(((...((....)).((.((((.......))))..(((((((((((((........)))))-------)))..)))))..)).)))....----- ( -21.80) >consensus UGGAAAAGCUGCGGCGGCGGAGCUUUCCACGCUCCCGAAGCGAAAAGCGUUCGCUCUCGCUUUCCACUUUUCACUUUUCUCGCCUCCGCCU_____ ..........((((.((((((((.......))))).((((.((((((.(...((....))....).)))))).))))....))).))))....... (-17.12 = -20.32 + 3.20)

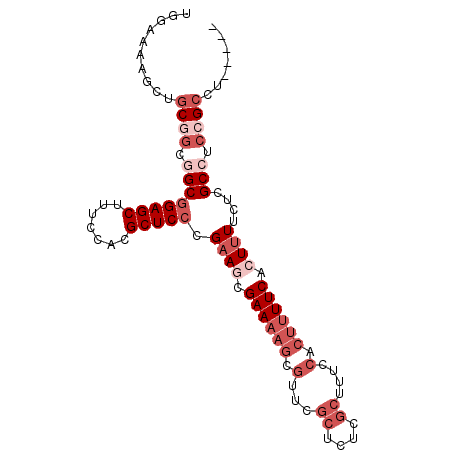
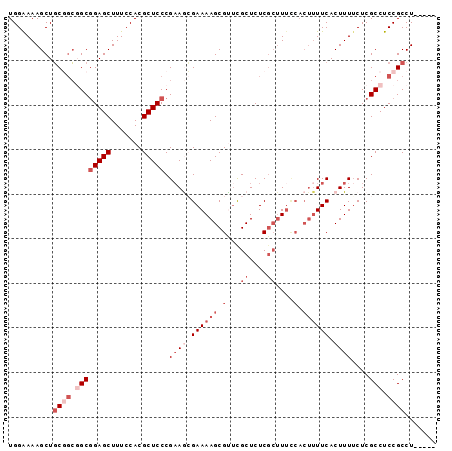
| Location | 19,327,812 – 19,327,903 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 86.02 |
| Mean single sequence MFE | -37.86 |
| Consensus MFE | -27.18 |
| Energy contribution | -28.98 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19327812 91 - 20766785 -----AGGCGGAGGCGAGAAAAGUGAAAAGUGAAAAGCGAGAGCGAACGCUUUUCGCUUCGGGAGCGUGGAAAGCUCCGCCGCCGCAGCUUUUCCA -----..((((.(((.....((((((((((((....((....))...))))))))))))..(((((.......)))))))).)))).......... ( -42.50) >DroSec_CAF1 5751 88 - 1 -----AGGCGGAGGCGAGCAAAGUGAAAAGUGGAA---GAGAGCGAGCGCUUUUCGCUUCGGGAGCGUGGAAAGCUCCGCCGCCGCAGCUUUUCCA -----..((((.(((...(.((((((((((((...---(....)...)))))))))))).)(((((.......)))))))).)))).......... ( -37.40) >DroSim_CAF1 5782 91 - 1 -----AGGCGGAGGCGAGAAAAGUGAAAAGUGGAAAGCGAGAGCGAGCGCUUUUCGCUUCGGGAGCGUGGAAAGCUCCGCCGCCGCAGCUUUCCCA -----..((((.(((.....((((((((((((....((....))...))))))))))))..(((((.......)))))))).)))).......... ( -42.50) >DroEre_CAF1 5836 96 - 1 GGCGGAGGCGGAGGCGAGAAAAGUGAAAAGUGGAAAGCGAGAGCGAACGCUUUUCUUUGCGGGAGCGUGGAAAGCUCCGCGCGGGAAGCUUUUCCA .((....))......(.(((((((((((((((....((....))...))))))))(((((((((((.......))))).))))))..)))))))). ( -37.60) >DroYak_CAF1 5799 84 - 1 -----AGGCGGAGGCGAGAAAAGUGAA-------AAGCGAGAGCAGACGCUUUUCUUUUCGGGAGCGUGGAAAGCUCAGCGGCAGCAGCUUUUCCA -----.((.((((((((((((((((..-------..((....))...)))))))))).(((.((((.......))))..))).....)))))))). ( -29.30) >consensus _____AGGCGGAGGCGAGAAAAGUGAAAAGUGGAAAGCGAGAGCGAACGCUUUUCGCUUCGGGAGCGUGGAAAGCUCCGCCGCCGCAGCUUUUCCA ......((.((((((.....((((((((((((....((....))...))))))))))))..(((((.......))))).........)))))))). (-27.18 = -28.98 + 1.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:42 2006