| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,291,278 – 19,291,372 |
| Length | 94 |
| Max. P | 0.754436 |

| Location | 19,291,278 – 19,291,372 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -17.82 |
| Consensus MFE | -10.48 |
| Energy contribution | -10.62 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19291278 94 + 20766785 UUUACCCACACAAAUCAUAGUGGGCCACAACUUCUGGUACAACAGUCCCAUUUACUUGACUCAACGCUCCCCUUUUCGAGCGUUUCCAUUUUUU------------- ....(((((..........)))))...(((....(((.((....)).))).....)))....(((((((........)))))))..........------------- ( -18.10) >DroSec_CAF1 41218 87 + 1 -------ACAGAAAUCAUAGUGGGCCACAACUUCUGGUACAACAGUCCCAUUUACUCGACUCAACGCUCCCCUUUUCGAGCGUUUCCAUUUUUU------------- -------...........(((((((((.......)))).....((((..........)))).(((((((........))))))).)))))....------------- ( -16.00) >DroSim_CAF1 31632 94 + 1 UUUACCCACACAAAUCAUAGUGGGCCACAACUUCUGGUACAACAGUCCCAUUUACUCGACUCAACGCUCCCCUUUUCGAGCGUUUCCAUUUUUU------------- ....(((((..........)))))..........(((......((((..........)))).(((((((........))))))).)))......------------- ( -18.10) >DroEre_CAF1 34080 94 + 1 UUUGCCCACACAAAUCAUUGUGGCCCACAACUUCUGGUACAACCGUCCCAUUUACUUGAUUUAACGCUCCCCUUUUCGAGCGUUUCCAUUUUUU------------- ...........(((((((((((...)))))....(((.((....)).)))......))))))(((((((........)))))))..........------------- ( -16.60) >DroYak_CAF1 31314 94 + 1 UUUGCCUACACAAAUCAUCGUGGCCCACAACUUCUGGUACAACAGUCCCAUUUACUUGAUUUAACGCUCCCCUUUUCGAGCGUUUCCAUUUUUU------------- ...........((((((..(((...)))......(((.((....)).)))......))))))(((((((........)))))))..........------------- ( -15.30) >DroAna_CAF1 39737 106 + 1 UUUGUUUGGCCAACCUUUCGGCGCCGACAACUUCUGGUACA-UAAUCCCAUUUACUUGAUUUAGCGCGCCCCUUUUUGGCAGUUUCCAGUUUUGAACUUCCAGCCAG ......((((.........(((((.(......((.((((.(-(......)).)))).)).....))))))......(((.(((((........))))).))))))). ( -22.80) >consensus UUUGCCCACACAAAUCAUAGUGGCCCACAACUUCUGGUACAACAGUCCCAUUUACUUGACUCAACGCUCCCCUUUUCGAGCGUUUCCAUUUUUU_____________ ........(((........)))............(((.((....)).)))............(((((((........)))))))....................... (-10.48 = -10.62 + 0.14)

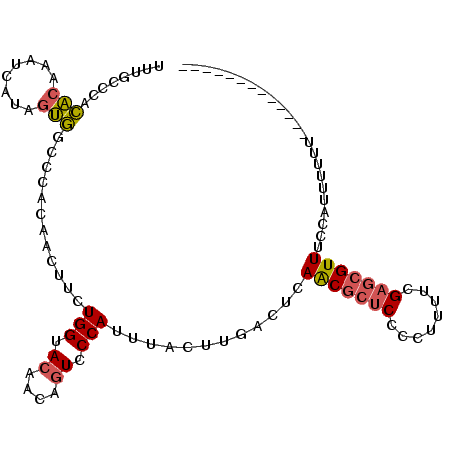

| Location | 19,291,278 – 19,291,372 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -14.30 |
| Energy contribution | -14.83 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19291278 94 - 20766785 -------------AAAAAAUGGAAACGCUCGAAAAGGGGAGCGUUGAGUCAAGUAAAUGGGACUGUUGUACCAGAAGUUGUGGCCCACUAUGAUUUGUGUGGGUAAA -------------......(((.(((((((........)))))))...)))......(((.((....)).))).........((((((..........))))))... ( -24.70) >DroSec_CAF1 41218 87 - 1 -------------AAAAAAUGGAAACGCUCGAAAAGGGGAGCGUUGAGUCGAGUAAAUGGGACUGUUGUACCAGAAGUUGUGGCCCACUAUGAUUUCUGU------- -------------..........(((((((........))))))).((((..........)))).......(((((((..(((....)))..))))))).------- ( -23.80) >DroSim_CAF1 31632 94 - 1 -------------AAAAAAUGGAAACGCUCGAAAAGGGGAGCGUUGAGUCGAGUAAAUGGGACUGUUGUACCAGAAGUUGUGGCCCACUAUGAUUUGUGUGGGUAAA -------------......(((.(((((((........))))))).((((..........))))......))).........((((((..........))))))... ( -24.80) >DroEre_CAF1 34080 94 - 1 -------------AAAAAAUGGAAACGCUCGAAAAGGGGAGCGUUAAAUCAAGUAAAUGGGACGGUUGUACCAGAAGUUGUGGGCCACAAUGAUUUGUGUGGGCAAA -------------..........(((((((........)))))))....(((.(...(((.((....)).)))..).)))..(.(((((........))))).)... ( -21.60) >DroYak_CAF1 31314 94 - 1 -------------AAAAAAUGGAAACGCUCGAAAAGGGGAGCGUUAAAUCAAGUAAAUGGGACUGUUGUACCAGAAGUUGUGGGCCACGAUGAUUUGUGUAGGCAAA -------------..........(((((((........)))))))....................((((..((.((((..(.(....).)..)))).))...)))). ( -19.70) >DroAna_CAF1 39737 106 - 1 CUGGCUGGAAGUUCAAAACUGGAAACUGCCAAAAAGGGGCGCGCUAAAUCAAGUAAAUGGGAUUA-UGUACCAGAAGUUGUCGGCGCCGAAAGGUUGGCCAAACAAA .((((..(...(((......(....).(((.......)))(((((....(((.(...(((.....-....)))..).)))..))))).)))...)..))))...... ( -28.30) >consensus _____________AAAAAAUGGAAACGCUCGAAAAGGGGAGCGUUAAAUCAAGUAAAUGGGACUGUUGUACCAGAAGUUGUGGCCCACUAUGAUUUGUGUGGGCAAA ...................(((.(((((((........)))))))...)))......(((.((....)).))).(((((((((....)))))))))........... (-14.30 = -14.83 + 0.53)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:32 2006