| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,285,510 – 19,285,624 |
| Length | 114 |
| Max. P | 0.751047 |

| Location | 19,285,510 – 19,285,624 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
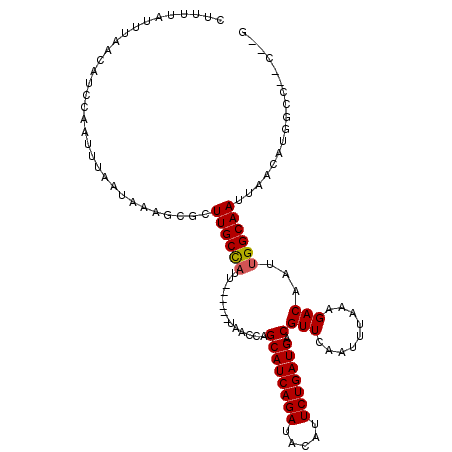
| Mean single sequence MFE | -18.19 |
| Consensus MFE | -14.73 |
| Energy contribution | -14.65 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
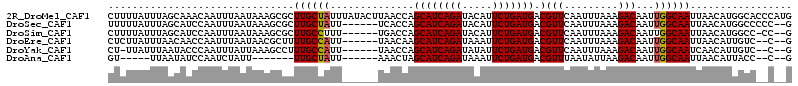
>2R_DroMel_CAF1 19285510 114 - 20766785 CUUUUAUUUAGCAAACAAUUUAAUAAAGCGCUUGCUAUUUAUACUUAACCAGCAUCAGAUACAUUCUGAUGACGUUCAAUUUAAAGACAAUUGGCAAUUAACAUGGCACCCAUG ....((..((((((.(.............).))))))..)).......(((((((((((.....)))))))..(((.........)))..)))).......(((((...))))) ( -18.72) >DroSec_CAF1 35426 106 - 1 UUUUUAUUUAGCAUCCAAUUUAAUAAAGCGCUUGCUAUU------UCACCAGCAUCAGAUACAUUCUGAUGACGUUCAAUUUAAAGACAAUUGGCAAUUAACAUGGCCCCC--G ........(((((..(.............)..)))))..------...(((((((((((.....))))))).)((.(((((.......)))))))........))).....--. ( -16.02) >DroSim_CAF1 25824 105 - 1 CUUUUAUUUAGCAUCCAAUUUAAUAAAGCGCUUGCCUUU------UGACCAGCAUCAGAUACAUUCUGAUGACGUUCAAUUUAAAGACAAUUGGCAAUUAACAUGGCC-CC--G ..............(((..(((((........((((..(------((....((((((((.....))))))).)..((........)))))..)))))))))..)))..-..--. ( -17.70) >DroEre_CAF1 28283 104 - 1 CUCUUAUUUAACAACCAAUUUAAUAACGCUUUUGCCAUU------UAACAAGCAUCAGAUAAAUUCUGAUGACGUUCAAUUUAAAGACAAUUGGCAAUUAACAUUGUC--C--G ...............................((((((..------......((((((((.....))))))).)(((.........)))...))))))...........--.--. ( -16.30) >DroYak_CAF1 25441 103 - 1 CU-UUAUUUAAUACCCAAUUUAUUAAAGCCUUUGCCAUU------UAACCAGCAUCAGAUAUAUUCUGAUGACGUUCAAUUUAAAGACAAUUGGCAAUCAACAUUGUC--C--G ..-...(((((((.......)))))))....((((((..------......((((((((.....))))))).)(((.........)))...))))))...........--.--. ( -17.60) >DroAna_CAF1 33953 92 - 1 GU-----UUAAUAUCCAAUCUAUU-------UUGCUAUU------AAACUAGCAUCAGAUAAAUUCUGAUGACGUUUAAUAUUAAGACAAUUGGCAAUUAACAUUACC--C--G ..-----(((((..(((((...((-------(((.((((------((((...(((((((.....)))))))..)))))))).)))))..)))))..))))).......--.--. ( -22.80) >consensus CUUUUAUUUAACAUCCAAUUUAAUAAAGCGCUUGCCAUU______UAACCAGCAUCAGAUACAUUCUGAUGACGUUCAAUUUAAAGACAAUUGGCAAUUAACAUGGCC__C__G ...............................((((((..............((((((((.....))))))).)(((.........)))...))))))................. (-14.73 = -14.65 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:30 2006