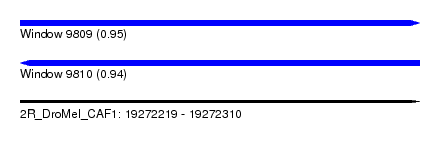
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,272,219 – 19,272,310 |
| Length | 91 |
| Max. P | 0.954431 |

| Location | 19,272,219 – 19,272,310 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -25.29 |
| Energy contribution | -24.85 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
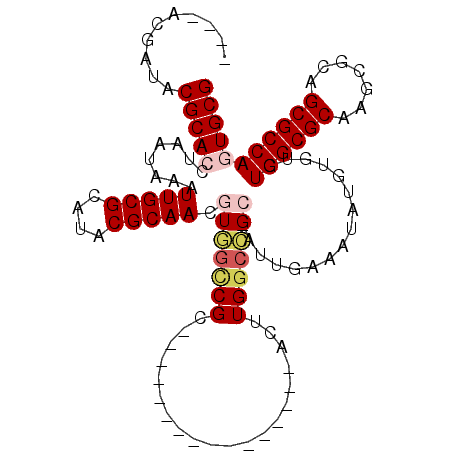
>2R_DroMel_CAF1 19272219 91 + 20766785 CGCAUUGGCGCUGCGCUUGCGCCACACACAUAUUUCAAU-GCGGCCAACU----------------GCGGCCACGUUGCGUAUGCGCAAUUUAUUAGGUGCGUAUCGU---- (((((((((((.......)))))...(.(((((..((((-(.((((....----------------..)))).))))).))))).)..........))))))......---- ( -33.10) >DroPse_CAF1 14944 112 + 1 CGCACUGGCGCUGCGCUUGCGCCACAAUCAUAUUUCAAUUUCAGCCAAGUGGCAGCUGCAGCGGUAGCGACUAUAUUGCGUAUACGCAAUUUAUUAGGUGCGGGUCGUGGUU .((....))((..((((((((((....((..............(((....))).(((((....))))))).((.((((((....)))))).))...)))))))).))..)). ( -39.70) >DroSim_CAF1 12656 91 + 1 CGCACUGGCGCUGCGCUUGCGCCACACACAUAUUUCAAU-GCGGCCAACU----------------GCGGCCACGUUGCGUAUGCGCAAUUUAUUAGGUGCGUAUCGU---- (((((((((((.......)))))...(.(((((..((((-(.((((....----------------..)))).))))).))))).)..........))))))......---- ( -35.50) >DroYak_CAF1 12024 81 + 1 CGCAUUGGCGCUGCGCUUGCGCCACAUACAUAUUUCAAU-GCG--------------------------GCCACGUUGCGUAUGCGCAAUUUAUUAGGUGCGUAUCGU---- ((((((.(((((((((.((.(((.(((..........))-).)--------------------------))))....))))).)))).........))))))......---- ( -25.80) >DroAna_CAF1 21384 92 + 1 CGCACUGGCGCUGCGCUUGCGCCACACACAUAUUUCAAU-GCAGCCAAAUG---------------GCGGCAACGUUGCGUAUACGCAAUUUAUUAGGUGCGUAUCGA---- (((((((((((.......)))))...............(-((.(((....)---------------)).)))..((((((....))))))......))))))......---- ( -33.60) >DroPer_CAF1 15439 112 + 1 CGCACUGGCGCUGCGCUUGCGCCACAAUCAUAUUUCAAUUUCAGCCAAGUGGCAGCUGCAGCGGUAGCGACUAUAUUGCGUAUACGCAAUUUAUUAGGUGCGGGUCGUGGUU .((....))((..((((((((((....((..............(((....))).(((((....))))))).((.((((((....)))))).))...)))))))).))..)). ( -39.70) >consensus CGCACUGGCGCUGCGCUUGCGCCACACACAUAUUUCAAU_GCAGCCAACU________________GCGGCCACGUUGCGUAUACGCAAUUUAUUAGGUGCGUAUCGU____ (((((((((((.......)))))...................................................((((((....))))))......)))))).......... (-25.29 = -24.85 + -0.44)

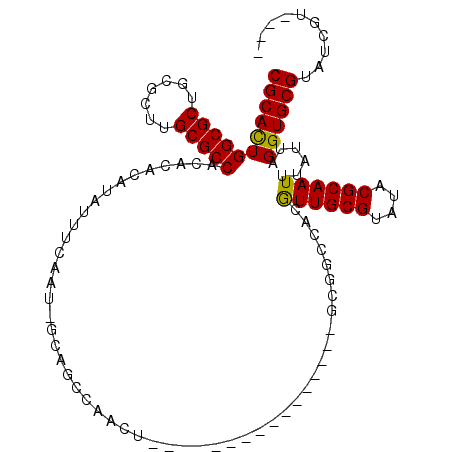
| Location | 19,272,219 – 19,272,310 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -32.78 |
| Consensus MFE | -23.58 |
| Energy contribution | -24.33 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19272219 91 - 20766785 ----ACGAUACGCACCUAAUAAAUUGCGCAUACGCAACGUGGCCGC----------------AGUUGGCCGC-AUUGAAAUAUGUGUGUGGCGCAAGCGCAGCGCCAAUGCG ----......((((...........(((((((..(((.(((((((.----------------...)))))))-.)))...))))))).((((((.......)))))).)))) ( -36.00) >DroPse_CAF1 14944 112 - 1 AACCACGACCCGCACCUAAUAAAUUGCGUAUACGCAAUAUAGUCGCUACCGCUGCAGCUGCCACUUGGCUGAAAUUGAAAUAUGAUUGUGGCGCAAGCGCAGCGCCAGUGCG ..........(((((.......((((((....))))))...........((((((....(((....))).....................((....))))))))...))))) ( -33.80) >DroSim_CAF1 12656 91 - 1 ----ACGAUACGCACCUAAUAAAUUGCGCAUACGCAACGUGGCCGC----------------AGUUGGCCGC-AUUGAAAUAUGUGUGUGGCGCAAGCGCAGCGCCAGUGCG ----......(((((..........(((((((..(((.(((((((.----------------...)))))))-.)))...))))))).((((((.......))))))))))) ( -38.90) >DroYak_CAF1 12024 81 - 1 ----ACGAUACGCACCUAAUAAAUUGCGCAUACGCAACGUGGC--------------------------CGC-AUUGAAAUAUGUAUGUGGCGCAAGCGCAGCGCCAAUGCG ----......((((.........(((((....)))))..((((--------------------------(((-((......)))).((((.(....)))))).)))).)))) ( -22.20) >DroAna_CAF1 21384 92 - 1 ----UCGAUACGCACCUAAUAAAUUGCGUAUACGCAACGUUGCCGC---------------CAUUUGGCUGC-AUUGAAAUAUGUGUGUGGCGCAAGCGCAGCGCCAGUGCG ----......(((((........(((((....)))))((.(((.((---------------(....))).))-).))...........((((((.......))))))))))) ( -32.00) >DroPer_CAF1 15439 112 - 1 AACCACGACCCGCACCUAAUAAAUUGCGUAUACGCAAUAUAGUCGCUACCGCUGCAGCUGCCACUUGGCUGAAAUUGAAAUAUGAUUGUGGCGCAAGCGCAGCGCCAGUGCG ..........(((((.......((((((....))))))...........((((((....(((....))).....................((....))))))))...))))) ( -33.80) >consensus ____ACGAUACGCACCUAAUAAAUUGCGCAUACGCAACGUGGCCGC________________ACUUGGCCGC_AUUGAAAUAUGUGUGUGGCGCAAGCGCAGCGCCAGUGCG ..........(((((........(((((....))))).(((((((....................)))))))................((((((.......))))))))))) (-23.58 = -24.33 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:25 2006