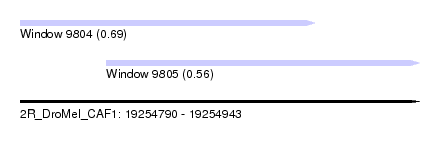
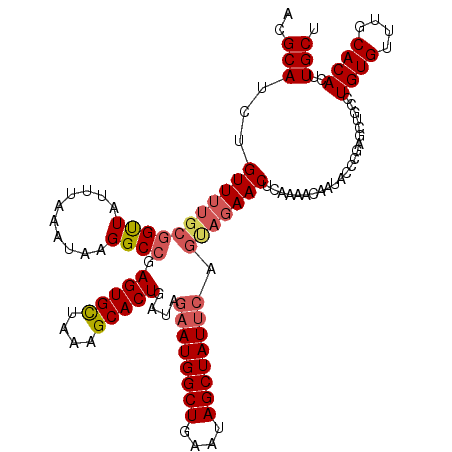
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,254,790 – 19,254,943 |
| Length | 153 |
| Max. P | 0.692230 |

| Location | 19,254,790 – 19,254,903 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 86.87 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -18.25 |
| Energy contribution | -19.50 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19254790 113 + 20766785 UUAUACAUCCUUCAAAAUCUCUAUGAACCAAACACGCAUCUGUUUUGCGGUUAUUUAAAUAAGGCCGAGUGCUAAAGCACUGCUAGAAUGGCUGAAUAGCUAUUCAGUAGAAC ...................(((((......(((.((((.......)))))))..........(((..(((((....)))))))).((((((((....)))))))).))))).. ( -27.40) >DroSec_CAF1 45471 113 + 1 UCAUACAUCCUUCAAAAUCUCUUUGAGCCAAACACGCAUCUGUUUUGCGGUUAUUUAAAUAAGGCCGAGUGCUAAAGCACUGAUAGAAUGGCUGAAUAGCUAUUCAGUAGAAC ((.((((((..(((((.....)))))(((.....((((.......)))).((((....)))))))..(((((....)))))))).((((((((....)))))))).))))).. ( -27.70) >DroSim_CAF1 44509 113 + 1 UCAUACAUCCUUCUAAAUCUCUUUGAGCCAAACACGCAUCUGUUUUGCGGUUAUUUAAAUAAGGCCGAGUGCUAAGGCACUGAUAGAAUGGCUGAAUAGCUAUUCAGCAGAAC ........(.(((((........((.(((.....((((.......)))).((((....)))))))))(((((....)))))..))))).)(((((((....)))))))..... ( -27.00) >DroYak_CAF1 45773 108 + 1 UGGAACAUCCUUCAAAAUAUCUAAGUGCCACAUACGCAUCUGUU-----GCUAUGUAAAUAAGGCAAAGUGUUAAAGCACUGAAAGAAUGGCUGAAUAGCUAUAGAAAAGAAC .((.....))(((......(((...(((((((((.(((.....)-----)))))))......)))).(((((....)))))...)))((((((....)))))).)))...... ( -22.60) >consensus UCAUACAUCCUUCAAAAUCUCUAUGAGCCAAACACGCAUCUGUUUUGCGGUUAUUUAAAUAAGGCCGAGUGCUAAAGCACUGAUAGAAUGGCUGAAUAGCUAUUCAGUAGAAC .........................................(((((((((((..........)))).(((((....)))))....((((((((....)))))))).))))))) (-18.25 = -19.50 + 1.25)

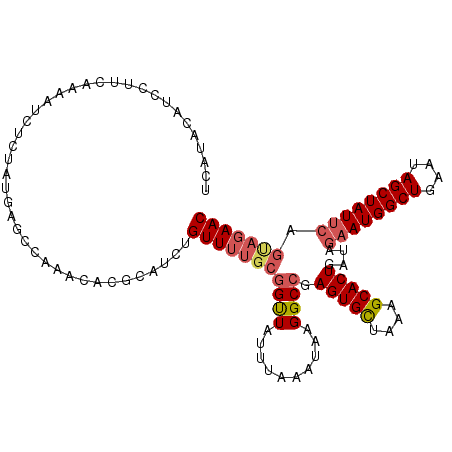
| Location | 19,254,823 – 19,254,943 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -25.38 |
| Energy contribution | -26.62 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19254823 120 + 20766785 ACGCAUCUGUUUUGCGGUUAUUUAAAUAAGGCCGAGUGCUAAAGCACUGCUAGAAUGGCUGAAUAGCUAUUCAGUAGAACUCAAAACAAUACCCGAGCUGCCUGUGUUUGCACACUUGCU ..(((..((((((..((((..........))))((((((....)).(((((.((((((((....))))))))))))).))))))))))..............((((....))))..))). ( -32.40) >DroSec_CAF1 45504 120 + 1 ACGCAUCUGUUUUGCGGUUAUUUAAAUAAGGCCGAGUGCUAAAGCACUGAUAGAAUGGCUGAAUAGCUAUUCAGUAGAACUGAAAACAAUACCCGAGCUGCCUGUGUUUGCACACUUGCU ..(((((.(((((((((((..........)))).(((((....)))))....((((((((....)))))))).))))))).))...................((((....))))..))). ( -32.10) >DroSim_CAF1 44542 120 + 1 ACGCAUCUGUUUUGCGGUUAUUUAAAUAAGGCCGAGUGCUAAGGCACUGAUAGAAUGGCUGAAUAGCUAUUCAGCAGAACUGAAAACAAUACCCGAGCUGCCUGUGUUUGCACACUUGCU ..(((((.(((((((((((..........)))).(((((....)))))....((((((((....)))))))).))))))).))...................((((....))))..))). ( -34.20) >DroYak_CAF1 45806 115 + 1 ACGCAUCUGUU-----GCUAUGUAAAUAAGGCAAAGUGUUAAAGCACUGAAAGAAUGGCUGAAUAGCUAUAGAAAAGAACACAAAACAAUAUCCGAGUUGCCUGUGUUUGCACACUUGCU ..(((..(((.-----.....((((((((((((((((((....)))))......((((((....))))))...........................)))))).))))))))))..))). ( -26.70) >consensus ACGCAUCUGUUUUGCGGUUAUUUAAAUAAGGCCGAGUGCUAAAGCACUGAUAGAAUGGCUGAAUAGCUAUUCAGUAGAACUCAAAACAAUACCCGAGCUGCCUGUGUUUGCACACUUGCU ..(((...(((((((((((..........)))).(((((....)))))....((((((((....)))))))).)))))))......................((((....))))..))). (-25.38 = -26.62 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:20 2006