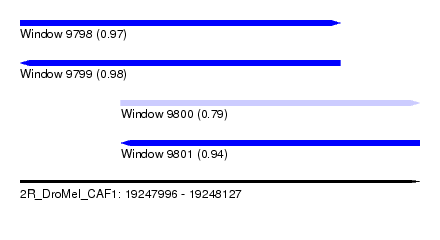
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,247,996 – 19,248,127 |
| Length | 131 |
| Max. P | 0.980471 |

| Location | 19,247,996 – 19,248,101 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -20.98 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19247996 105 + 20766785 UCAAAUAUA----UAGACAACUCGGCCACUUGGCGGGCUAAUAUUUGAAUAUUUGAAUAUUUUGUGUUUGUAUUUGUCUGCGGCUGU-------GUGCUUAGCCGCAAUUCUUGGC .((((((((----.(.((((((((.((....))))))..((((((..(....)..)))))))))).).))))))))..((((((((.-------.....))))))))......... ( -31.60) >DroSec_CAF1 38577 105 + 1 UCAAAUAUA----UAGACAACUCGGCCACUUGGCGGGCUAAUAUUUGAAUAUUUGAAUAUUUUGUGUUUGUAUUUGUCUGCGGCUGU-------GUGCUUAGCCGCAAUUCUUGGC .((((((((----.(.((((((((.((....))))))..((((((..(....)..)))))))))).).))))))))..((((((((.-------.....))))))))......... ( -31.60) >DroSim_CAF1 37225 105 + 1 UCAAAUAUA----UAGACAACUCGGCCACUUGGCGGGCUAAUAUUUGAAUAUUUGAAUAUUUUGUGUUUGUAUUUGCCUGCGGCUGU-------GUGCUUAGCCGCAAUUCUUGGC .((((((((----.(.((((((((.((....))))))..((((((..(....)..)))))))))).).))))))))..((((((((.-------.....))))))))......... ( -31.40) >DroEre_CAF1 38519 86 + 1 UCAAAUAUACGUGUAUACCACUUGGCCACUUGGCGGGCUAAUAUU-----------------------UGUAUUUGUCUGCGGCUGU-------GUGCUUAGCCGCAAUUCUUGGC .((((((((.(((.....)))((((((........))))))....-----------------------))))))))..((((((((.-------.....))))))))......... ( -26.70) >DroYak_CAF1 38765 93 + 1 UCAAAUAU------AUACAACUUGGCCACUUGGCGGGCUAAUAUUUGGA-----------------UUUGUAUUUGUCUGCGGCUGUGUGCUGUGUGCUUAGCCGCAAUUCUUGGC .(((((((------(..(((.((((((........))))))...)))..-----------------..))))))))..((((((((.(..(...)..).))))))))......... ( -29.10) >consensus UCAAAUAUA____UAGACAACUCGGCCACUUGGCGGGCUAAUAUUUGAAUAUUUGAAUAUUUUGUGUUUGUAUUUGUCUGCGGCUGU_______GUGCUUAGCCGCAAUUCUUGGC ........................((((....((((((.(((((..((((((...........))))))))))).))))))(((((.............)))))........)))) (-20.98 = -21.30 + 0.32)

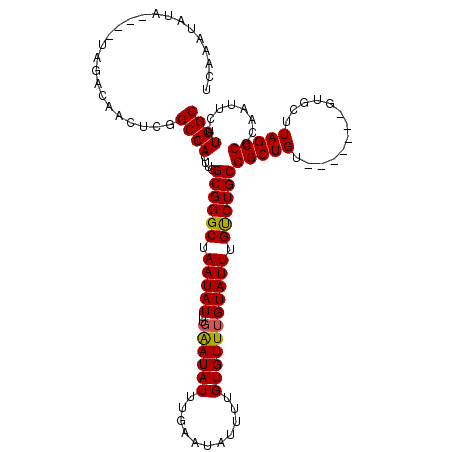
| Location | 19,247,996 – 19,248,101 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -24.13 |
| Consensus MFE | -16.94 |
| Energy contribution | -17.54 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19247996 105 - 20766785 GCCAAGAAUUGCGGCUAAGCAC-------ACAGCCGCAGACAAAUACAAACACAAAAUAUUCAAAUAUUCAAAUAUUAGCCCGCCAAGUGGCCGAGUUGUCUA----UAUAUUUGA ........((((((((......-------..)))))))).....................((((((((........(((((.(((....))).).))))....----.)))))))) ( -23.92) >DroSec_CAF1 38577 105 - 1 GCCAAGAAUUGCGGCUAAGCAC-------ACAGCCGCAGACAAAUACAAACACAAAAUAUUCAAAUAUUCAAAUAUUAGCCCGCCAAGUGGCCGAGUUGUCUA----UAUAUUUGA ........((((((((......-------..)))))))).....................((((((((........(((((.(((....))).).))))....----.)))))))) ( -23.92) >DroSim_CAF1 37225 105 - 1 GCCAAGAAUUGCGGCUAAGCAC-------ACAGCCGCAGGCAAAUACAAACACAAAAUAUUCAAAUAUUCAAAUAUUAGCCCGCCAAGUGGCCGAGUUGUCUA----UAUAUUUGA (((......(((((((......-------..))))))))))...................((((((((........(((((.(((....))).).))))....----.)))))))) ( -26.22) >DroEre_CAF1 38519 86 - 1 GCCAAGAAUUGCGGCUAAGCAC-------ACAGCCGCAGACAAAUACA-----------------------AAUAUUAGCCCGCCAAGUGGCCAAGUGGUAUACACGUAUAUUUGA ........((((((((......-------..)))))))).......((-----------------------(((((..((..(((....)))...(((.....)))))))))))). ( -23.80) >DroYak_CAF1 38765 93 - 1 GCCAAGAAUUGCGGCUAAGCACACAGCACACAGCCGCAGACAAAUACAAA-----------------UCCAAAUAUUAGCCCGCCAAGUGGCCAAGUUGUAU------AUAUUUGA ........((((((((..((.....))....))))))))...........-----------------..(((((((((((..(((....)))...))))...------))))))). ( -22.80) >consensus GCCAAGAAUUGCGGCUAAGCAC_______ACAGCCGCAGACAAAUACAAACACAAAAUAUUCAAAUAUUCAAAUAUUAGCCCGCCAAGUGGCCGAGUUGUCUA____UAUAUUUGA ........((((((((...............))))))))..............................(((((((((((..(((....)))...)))).........))))))). (-16.94 = -17.54 + 0.60)

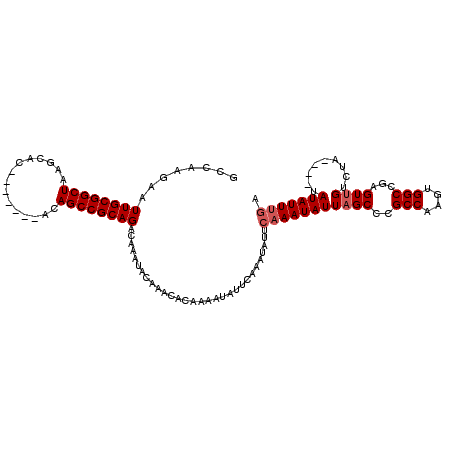
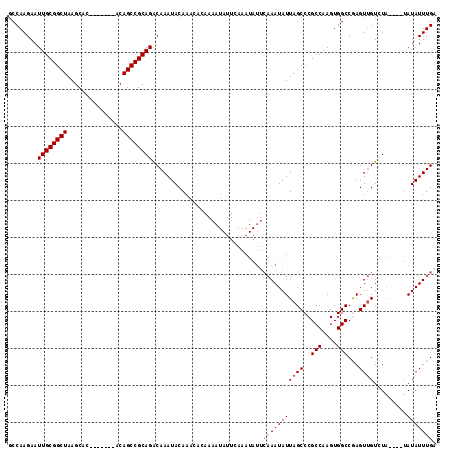
| Location | 19,248,029 – 19,248,127 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.74 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -18.68 |
| Energy contribution | -19.16 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19248029 98 + 20766785 CUAAUAUUUGAAUAUUUGAAUAUUUUGUGUUUGUAUUUGUCUGCGGCUGU-------GUGCUUAGCCGCAAUUCUUGGCAAGCAAACUGCUUUGGCAACACCCGC ..((((((..(....)..)))))).(((((((((..(((((((((((((.-------.....))))))))......)))))))))))(((....))))))..... ( -26.70) >DroSec_CAF1 38610 98 + 1 CUAAUAUUUGAAUAUUUGAAUAUUUUGUGUUUGUAUUUGUCUGCGGCUGU-------GUGCUUAGCCGCAAUUCUUGGCAAGCAAACUGUUUUGGCAACACCCGC ..((((((..(....)..))))))....((((((..(((((((((((((.-------.....))))))))......))))))))))).....((....))..... ( -25.80) >DroSim_CAF1 37258 98 + 1 CUAAUAUUUGAAUAUUUGAAUAUUUUGUGUUUGUAUUUGCCUGCGGCUGU-------GUGCUUAGCCGCAAUUCUUGGCAAGCAAACUGUUUUGGCAACACCCGC ..((((((..(....)..))))))....((((((..(((((((((((((.-------.....))))))))......))))))))))).....((....))..... ( -28.50) >DroEre_CAF1 38556 75 + 1 CUAAUAUU-----------------------UGUAUUUGUCUGCGGCUGU-------GUGCUUAGCCGCAAUUCUUGGCAAGCAAACUGUUUUGGCAGCACCCGC ......((-----------------------(((..(((((((((((((.-------.....))))))))......))))))))))((((....))))....... ( -21.00) >DroYak_CAF1 38796 88 + 1 CUAAUAUUUGGA-----------------UUUGUAUUUGUCUGCGGCUGUGUGCUGUGUGCUUAGCCGCAAUUCUUGGCAAGCAAACUGUUUUGGCAGCACCCGC .........(((-----------------(........))))((((....(((((((.(((((.((((.......)))))))))..(......)))))))))))) ( -25.50) >consensus CUAAUAUUUGAAUAUUUGAAUAUUUUGUGUUUGUAUUUGUCUGCGGCUGU_______GUGCUUAGCCGCAAUUCUUGGCAAGCAAACUGUUUUGGCAACACCCGC ............................((((((..(((((((((((((.............))))))))......)))))))))))(((....)))........ (-18.68 = -19.16 + 0.48)

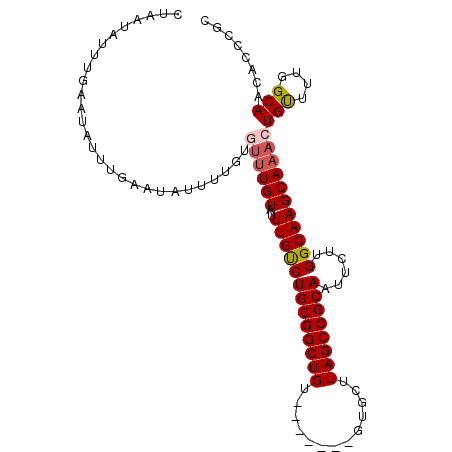
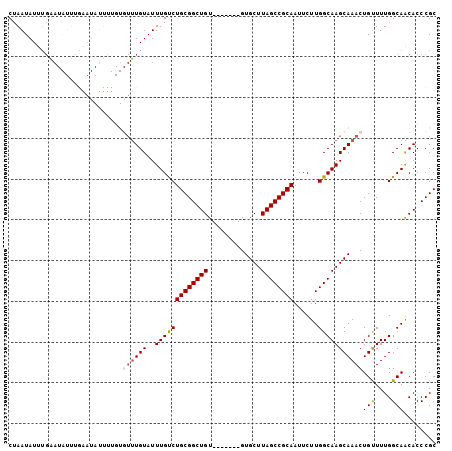
| Location | 19,248,029 – 19,248,127 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 82.74 |
| Mean single sequence MFE | -21.94 |
| Consensus MFE | -19.12 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19248029 98 - 20766785 GCGGGUGUUGCCAAAGCAGUUUGCUUGCCAAGAAUUGCGGCUAAGCAC-------ACAGCCGCAGACAAAUACAAACACAAAAUAUUCAAAUAUUCAAAUAUUAG ((((.((((((....)))...((((((((.........))).))))).-------))).)))).......................................... ( -21.60) >DroSec_CAF1 38610 98 - 1 GCGGGUGUUGCCAAAACAGUUUGCUUGCCAAGAAUUGCGGCUAAGCAC-------ACAGCCGCAGACAAAUACAAACACAAAAUAUUCAAAUAUUCAAAUAUUAG ((((.((((.....))))((.((((((((.........))).))))).-------))..)))).......................................... ( -20.20) >DroSim_CAF1 37258 98 - 1 GCGGGUGUUGCCAAAACAGUUUGCUUGCCAAGAAUUGCGGCUAAGCAC-------ACAGCCGCAGGCAAAUACAAACACAAAAUAUUCAAAUAUUCAAAUAUUAG ..(((((((.........(((((.(((((......(((((((......-------..))))))))))))...)))))............)))))))......... ( -23.20) >DroEre_CAF1 38556 75 - 1 GCGGGUGCUGCCAAAACAGUUUGCUUGCCAAGAAUUGCGGCUAAGCAC-------ACAGCCGCAGACAAAUACA-----------------------AAUAUUAG ((((((((((......))))..))))))......((((((((......-------..)))))))).........-----------------------........ ( -21.90) >DroYak_CAF1 38796 88 - 1 GCGGGUGCUGCCAAAACAGUUUGCUUGCCAAGAAUUGCGGCUAAGCACACAGCACACAGCCGCAGACAAAUACAAA-----------------UCCAAAUAUUAG ((((((((((......))))..))))))......((((((((..((.....))....))))))))...........-----------------............ ( -22.80) >consensus GCGGGUGUUGCCAAAACAGUUUGCUUGCCAAGAAUUGCGGCUAAGCAC_______ACAGCCGCAGACAAAUACAAACACAAAAUAUUCAAAUAUUCAAAUAUUAG ((((.(((((......)))..((((((((.........))).))))).........)).)))).......................................... (-19.12 = -18.88 + -0.24)

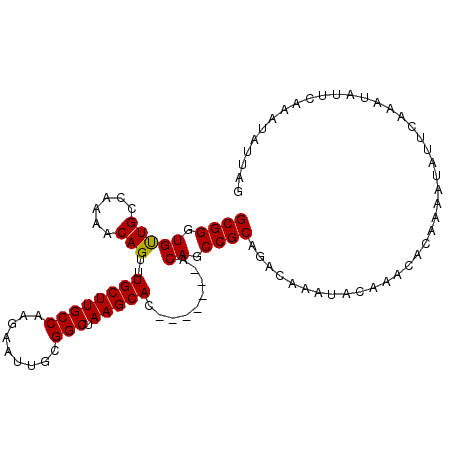
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:16 2006