| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,243,871 – 19,243,981 |
| Length | 110 |
| Max. P | 0.809915 |

| Location | 19,243,871 – 19,243,981 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.15 |
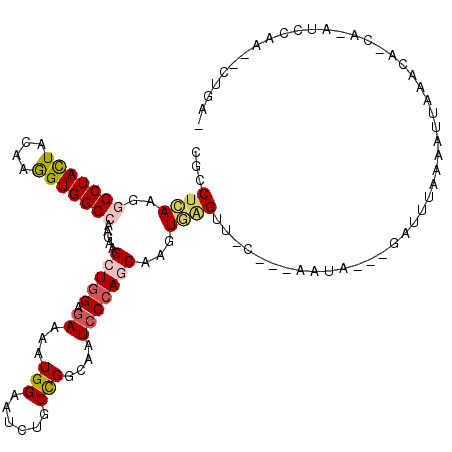
| Mean single sequence MFE | -28.79 |
| Consensus MFE | -17.09 |
| Energy contribution | -17.53 |
| Covariance contribution | 0.45 |
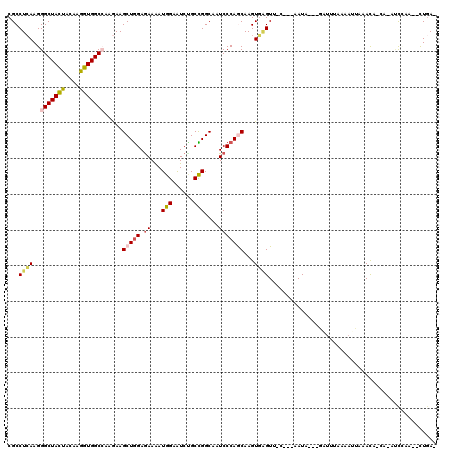
| Combinations/Pair | 1.24 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
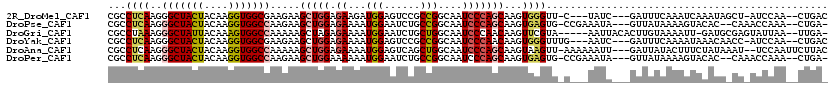
>2R_DroMel_CAF1 19243871 110 - 20766785 CGCCUCAAGGGCUACUACAAGGUGGCGAAGAAGCUGGAGAAGAUGGAGUCCGCCGGCAAUCCCAGCAAGUGGGUU-C---UAUC---GAUUUCAAAUCAAAUAGCU-AUCCAA--CUGAC ........((((((((....)))))).....(((((.....((((((..((((..((.......))..))))..)-)---))))---(((.....)))...)))))-..))..--..... ( -33.70) >DroPse_CAF1 36970 111 - 1 CGCCUCAAGGGCUACUACAAGGUGGCCAAGAAGCUGGAGAAAAUGGAAUCUGCCGGCAAUCCCAGCAAGUGAGUG-CCGAAAUA---GUUAUAAAAGUACAC--CAAACCAAA--CUGA- .((((((..(((((((....))))))).....(((((.((...(((......)))....)))))))...)))).)-).....((---(((............--.......))--))).- ( -29.61) >DroGri_CAF1 44797 111 - 1 CGCCUAAAGGGCUAUUACAAAGUGGCCAAAAAGCUAGAGAAAAUGGAAUCUGCUGGCAAUCCCAACAAGUUCGUA-----AAUUACACUUGUAAAAUU-GAUGCGAGUAUUAA--UUGA- .(((.....(((((((....)))))))....(((.(((..........))))))))).......((((((..(((-----...)))))))))..((((-((((....))))))--))..- ( -24.80) >DroYak_CAF1 34631 111 - 1 CGCCUCAAGGGCUACUACAAGGUGGCGAAGAAGCUGGAGAAAAUGGAGUCCGCCGGCAAUCCCAACAAGUGGGUUUG---AAUC---GAUUUCAAAAUAAACAACC-AUCCAA--CUGAC .(((.....)))........(((((.......(((((.((........))..)))))...((((.....))))((((---((..---...))))))........))-)))...--..... ( -24.50) >DroAna_CAF1 33834 114 - 1 CGCCUCAAGGGCUACUACAAGGUGGCCAAAAAGCUGGAGAAAAUGGAGUCAGCUGGCAAUCCCAGCAAGUAAGUU-AAAAAAUU---GAUUAUACUUUCUAUAAAU--UCCAAUUCUUAC .((......(((((((....))))))).....))..(((((..((((((..(((((.....)))))(((((((((-((....))---)))).))))).......))--)))).))))).. ( -31.70) >DroPer_CAF1 36032 111 - 1 CGCCUCAAGGGCUACUACAAGGUGGCCAAGAAGCUGGAAAAAAUGGAAUCUGCCGGCAAUCCCAGCAAGUGAGUG-CCGAAAUA---GUUAUAAAAGUACAC--CAAACCAAA--CUGA- .((((((..(((((((....))))))).....(((((......(((......)))......)))))...)))).)-).....((---(((............--.......))--))).- ( -28.41) >consensus CGCCUCAAGGGCUACUACAAGGUGGCCAAGAAGCUGGAGAAAAUGGAAUCUGCCGGCAAUCCCAGCAAGUGAGUU_C___AAUA___GAUUUAAAAUUAAACA_CA_AUCCAA__CUGA_ ...((((..(((((((....))))))).....(((((.((...(((......)))....)))))))...))))............................................... (-17.09 = -17.53 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:12 2006