| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,241,631 – 19,241,730 |
| Length | 99 |
| Max. P | 0.979119 |

| Location | 19,241,631 – 19,241,730 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.54 |
| Mean single sequence MFE | -20.14 |
| Consensus MFE | -12.19 |
| Energy contribution | -11.41 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
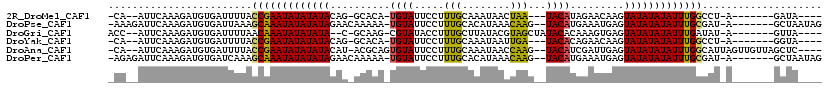
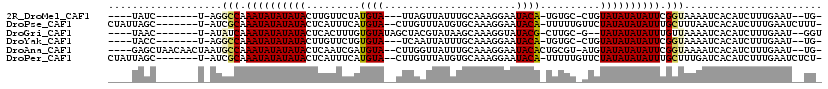
>2R_DroMel_CAF1 19241631 99 + 20766785 -CA--AUUCAAAGAUGUGAUUUUACCGAAUAUAUAUACAG-GCACA-UGUAUUCCUUUGCAAAUAACUAA---UACAUAGAACAAGUAUAUAUAUUUGGCCU-A-------GAUA---- -..--.............((((..((((((((((((((..-(..((-((((((..((.......))..))---))))).)..)..))))))))))))))...-)-------))).---- ( -19.10) >DroPse_CAF1 33988 107 + 1 -AAAGAUUCAAAGAUGUGAUUAAAGCAAAUAUAUAUAGAACAAAAA-UGUAUUCCUUUGCACAUAAACAAG--UACAUGAAAUGAGUAUAUAUAUUUGCGAU-A-------GCUAAUAG -.......................(((((((((((((........(-((((((..((((....))))..))--)))))........)))))))))))))...-.-------........ ( -19.49) >DroGri_CAF1 42396 101 + 1 ACC--AUUCAAAGAUGUGAUUUUAACAAAUAUAUAUA--C-GCAAG-CGUAUACCUUUGCUUAUACGUAGCUAUACACAAAGUGAGUAUAUAUAUUUGAUAU-A-------GUUA---- ...--...........(((((.((.((((((((((((--(-((..(-((((((........))))))).))....(((...))).))))))))))))).)).-)-------))))---- ( -23.00) >DroYak_CAF1 32349 99 + 1 -CA--AUUCAAAGAUGUGAUUUUACCGAAUAUAUAUACAG-GCACA-UGUAUUCCUUUGCAAAUAAUUGA---UACACAGAACAAGUAUAUAUAUUUGGCCU-A-------GGUA---- -..--...................((((((((((((((..-.....-((((......)))).....(((.---....))).....))))))))))))))...-.-------....---- ( -17.60) >DroAna_CAF1 31568 109 + 1 -CA--AUUCAAAGAUGUGAUUUUACCGAAUAUAUAUACAU-ACGCAGUGUAUUCCUUUGCAAAUAACCAAG--UACAUCGAUUGAGUAUAUAUAUUUGGCAUUAGUUGUUAGCUC---- -..--......((..(..(((...((((((((((((((..-...(.(((((((................))--))))).).....))))))))))))))....)))..)...)).---- ( -20.99) >DroPer_CAF1 33063 107 + 1 -AGAGAUUCAAAGAUGUGAUCAAAGCAAAUAUAUAUAGAACAAAAA-UGUAUUCCUUUGCACAUAAACAAG--UACAUGAAAUGAGUAUAUAUAUUUGCGAU-A-------GCUAAUAG -...((((((....)).))))...(((((((((((((........(-((((((..((((....))))..))--)))))........)))))))))))))...-.-------........ ( -20.69) >consensus _CA__AUUCAAAGAUGUGAUUUUACCAAAUAUAUAUACAA_ACAAA_UGUAUUCCUUUGCAAAUAAACAAG__UACAUAAAAUGAGUAUAUAUAUUUGGCAU_A_______GCUA____ ........................(((((((((((((..........((((.....(((........)))...)))).........))))))))))))).................... (-12.19 = -11.41 + -0.78)
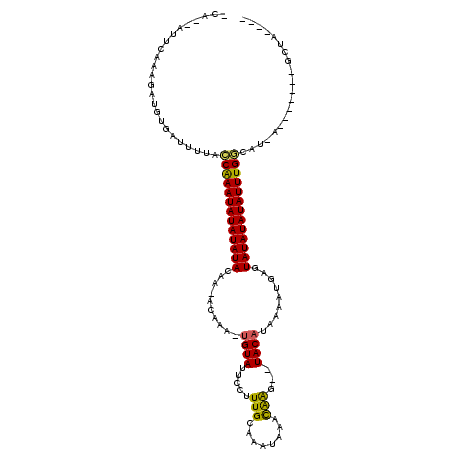
| Location | 19,241,631 – 19,241,730 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.54 |
| Mean single sequence MFE | -20.97 |
| Consensus MFE | -7.45 |
| Energy contribution | -7.39 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
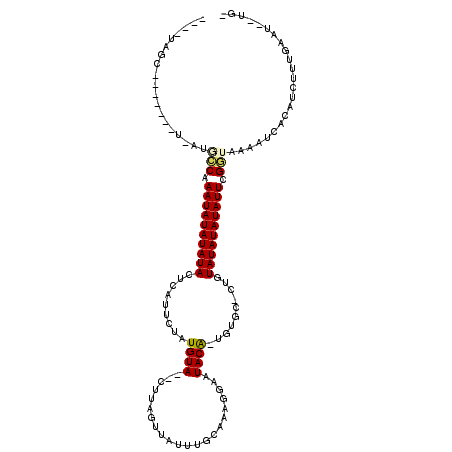
>2R_DroMel_CAF1 19241631 99 - 20766785 ----UAUC-------U-AGGCCAAAUAUAUAUACUUGUUCUAUGUA---UUAGUUAUUUGCAAAGGAAUACA-UGUGC-CUGUAUAUAUAUUCGGUAAAAUCACAUCUUUGAAU--UG- ----....-------.-..(((.(((((((((((..((..((((((---((..((.......))..))))))-)).))-..))))))))))).)))..................--..- ( -21.00) >DroPse_CAF1 33988 107 - 1 CUAUUAGC-------U-AUCGCAAAUAUAUAUACUCAUUUCAUGUA--CUUGUUUAUGUGCAAAGGAAUACA-UUUUUGUUCUAUAUAUAUUUGCUUUAAUCACAUCUUUGAAUCUUU- ........-------.-...((((((((((((..........((((--(........)))))..((((((..-....)))))))))))))))))).......................- ( -20.80) >DroGri_CAF1 42396 101 - 1 ----UAAC-------U-AUAUCAAAUAUAUAUACUCACUUUGUGUAUAGCUACGUAUAAGCAAAGGUAUACG-CUUGC-G--UAUAUAUAUUUGUUAAAAUCACAUCUUUGAAU--GGU ----..((-------(-((..(((((((((((((.((....(((((((.((..((....))..)).))))))-).)).-)--))))))))))))......(((......)))))--))) ( -25.10) >DroYak_CAF1 32349 99 - 1 ----UACC-------U-AGGCCAAAUAUAUAUACUUGUUCUGUGUA---UCAAUUAUUUGCAAAGGAAUACA-UGUGC-CUGUAUAUAUAUUCGGUAAAAUCACAUCUUUGAAU--UG- ----....-------.-..(((.(((((((((((..((...(((((---((((....))).......)))))-)..))-..))))))))))).)))..................--..- ( -18.51) >DroAna_CAF1 31568 109 - 1 ----GAGCUAACAACUAAUGCCAAAUAUAUAUACUCAAUCGAUGUA--CUUGGUUAUUUGCAAAGGAAUACACUGCGU-AUGUAUAUAUAUUCGGUAAAAUCACAUCUUUGAAU--UG- ----..............((((.(((((((((((..((((((....--.))))))....(((..(.....)..)))..-..))))))))))).)))).................--..- ( -19.40) >DroPer_CAF1 33063 107 - 1 CUAUUAGC-------U-AUCGCAAAUAUAUAUACUCAUUUCAUGUA--CUUGUUUAUGUGCAAAGGAAUACA-UUUUUGUUCUAUAUAUAUUUGCUUUGAUCACAUCUUUGAAUCUCU- ........-------.-...((((((((((((..........((((--(........)))))..((((((..-....))))))))))))))))))...(((((......)).)))...- ( -21.00) >consensus ____UAGC_______U_AUGCCAAAUAUAUAUACUCAUUCUAUGUA__CUUAGUUAUUUGCAAAGGAAUACA_UGUGC_CUGUAUAUAUAUUCGGUAAAAUCACAUCUUUGAAU__UG_ ...................(((.((((((((((.........((((......................))))..........)))))))))).)))....................... ( -7.45 = -7.39 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:10 2006