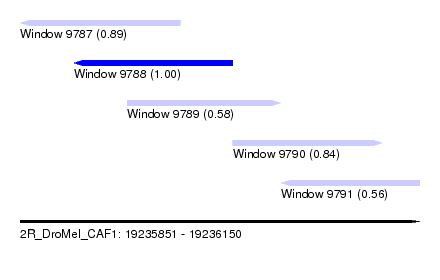
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,235,851 – 19,236,150 |
| Length | 299 |
| Max. P | 0.999902 |

| Location | 19,235,851 – 19,235,971 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.16 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -19.19 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
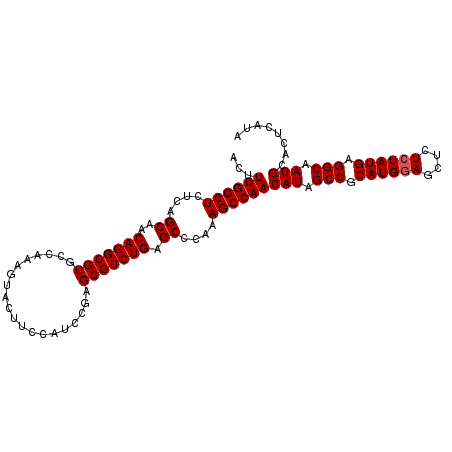
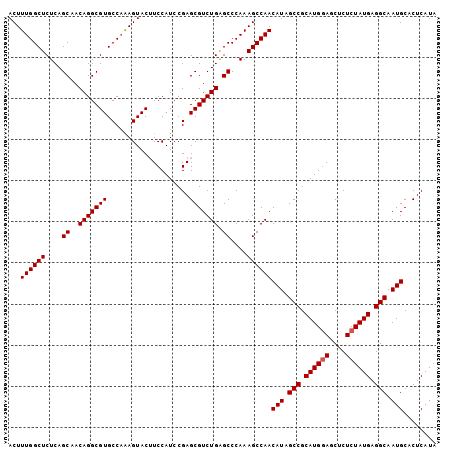
>2R_DroMel_CAF1 19235851 120 - 20766785 UUCUGGCACUCGCUUCCCGCCACCAAUGGCUGGCUGGUGGCCAUGGCUACCGUGGCUCGCAGCCCCCUUUUCAAUAAAGAUUUUAGCUGCAAUCAGGACCUUUUUGCAUCUGCAGGUGCA .....((((..((..((.((((....)))).))(((((((((((((...)))))))).(((((...((((.....))))......))))).)))))...............))..)))). ( -41.00) >DroSim_CAF1 25285 110 - 1 UUCUGGCACUCGGCUCCC-------AUGGCU--CUGGUGGCCAUGGCUACCGUGGCUCGCAGCCCCCCUUUCAAUAAAGAUUUGAGCUGCAAUCAGGAUAU-UUUGCAUCUGCAGGUGCA (((((((.(.(((...((-------((((((--.....))))))))...))).)))..(((((.......((((.......)))))))))...)))))...-..((((((....)))))) ( -38.61) >DroEre_CAF1 26385 101 - 1 UUCUGUUC--------CCACCAUCAAUG--------GUGGCCAUGGCUACUGGGGCUCGCAGCCACCUUU--AGUAAAGAUUUAAGCUGCAAUCAGUAUAU-UUCGCAUGGGCAGGUGCG .(((((..--------((((((....))--------))))(((((((((..(((((.....))).))..)--)))...((..((.((((....)))).)).-.)).)))))))))).... ( -35.40) >DroYak_CAF1 26456 91 - 1 UUCUGGCA------------CAUCAAUGGC----CGGUGGUCAUGGCUACUGUG----GCAGCCACCUGU--AAUAAGGAUUUAUGCUGCAAUCAAGAUGC-UUUG------CAGCUGCA ...((((.------------((....))((----(((((((....))))))..)----)).))))(((..--....)))......(((((((.........-.)))------)))).... ( -28.70) >consensus UUCUGGCA________CC__CAUCAAUGGC____UGGUGGCCAUGGCUACCGUGGCUCGCAGCCACCUUU__AAUAAAGAUUUAAGCUGCAAUCAGGAUAU_UUUGCAUCUGCAGGUGCA (((((.............................(((((((....)))))))......(((((......................)))))...)))))......(((((......))))) (-19.19 = -20.12 + 0.94)

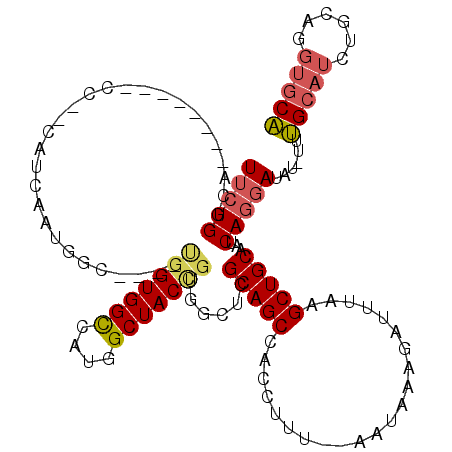
| Location | 19,235,891 – 19,236,010 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.83 |
| Mean single sequence MFE | -39.30 |
| Consensus MFE | -26.40 |
| Energy contribution | -25.63 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.67 |
| SVM decision value | 4.46 |
| SVM RNA-class probability | 0.999902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19235891 119 - 20766785 UACAACAA-UUGCUUGAUGUUCUGCCGAGCUCUGCCAGAUUUCUGGCACUCGCUUCCCGCCACCAAUGGCUGGCUGGUGGCCAUGGCUACCGUGGCUCGCAGCCCCCUUUUCAAUAAAGA .......(-(((......(..((((.((((..((((((....))))))..(((..((.((((....)))).))..((((((....)))))))))))))))))..)......))))..... ( -42.10) >DroSim_CAF1 25324 110 - 1 UGCGGCAA-UUGCUUGAUGCUCUGCCGAGCUCUGCCAGAUUUCUGGCACUCGGCUCCC-------AUGGCU--CUGGUGGCCAUGGCUACCGUGGCUCGCAGCCCCCCUUUCAAUAAAGA (((((((.-..((.....))..))))((((..((((((....))))))(.(((...((-------((((((--.....))))))))...))).))))))))................... ( -45.20) >DroYak_CAF1 26489 92 - 1 ---AGCAAUUUGCUUGAUGUU---CCGAGUUUUGCUGGAUUUCUGGCA------------CAUCAAUGGC----CGGUGGUCAUGGCUACUGUG----GCAGCCACCUGU--AAUAAGGA ---(((((...(((((.....---.))))).))))).(((..(((((.------------((....))))----)))..))).(((((.(....----).)))))(((..--....))). ( -30.60) >consensus U_CAGCAA_UUGCUUGAUGUUCUGCCGAGCUCUGCCAGAUUUCUGGCACUCG__UCCC__CA_CAAUGGCU__CUGGUGGCCAUGGCUACCGUGGCUCGCAGCCCCCUUUUCAAUAAAGA ...........(((((.........)))))..((((((....))))))...................((((....((((((....))))))((.....))))))................ (-26.40 = -25.63 + -0.77)

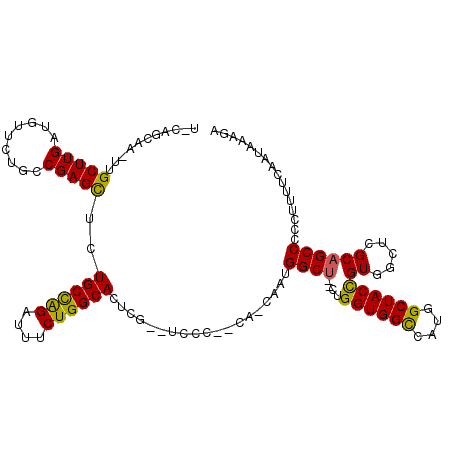
| Location | 19,235,931 – 19,236,046 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -17.99 |
| Energy contribution | -19.33 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19235931 115 + 20766785 CCACCAGCCAGCCAUUGGUGGCGGGAAGCGAGUGCCAGAAAUCUGGCAGAGCUCGGCAGAACAUCAAGCAA-UUGUUGUACCCGUUGGUCCGAGUUUAUUAAAUUCCGCAUUUCAU .((((((.......))))))((((((......((((((....))))))((((((((....(((.(((....-))).)))(((....)))))))))))......))))))....... ( -41.00) >DroSim_CAF1 25364 106 + 1 CCACCAG--AGCCAU-------GGGAGCCGAGUGCCAGAAAUCUGGCAGAGCUCGGCAGAGCAUCAAGCAA-UUGCCGCACCCGUUGGUCCGAGUUUAUUAAAUUCCGCAUUUCAU ......(--..((((-------(((.((((((((((((....)))))...)))))))...((..(((....-)))..)).)))).)))..)(((((.....))))).......... ( -38.50) >DroYak_CAF1 26523 93 + 1 CCACCG----GCCAUUGAUG------------UGCCAGAAAUCCAGCAAAACUCGG---AACAUCAAGCAAAUUGCU----CCGUAGGUCCGAACUUAUUAAAUUCCGCAUUUCAA ....((----(((.((((((------------(.((((.............)).))---.)))))))((.....)).----.....)).)))........................ ( -16.62) >consensus CCACCAG__AGCCAUUG_UG__GGGA__CGAGUGCCAGAAAUCUGGCAGAGCUCGGCAGAACAUCAAGCAA_UUGCUG_ACCCGUUGGUCCGAGUUUAUUAAAUUCCGCAUUUCAU ..........((....................((((((....))))))((((((((...(((....(((.....)))......)))...))))))))..........))....... (-17.99 = -19.33 + 1.34)

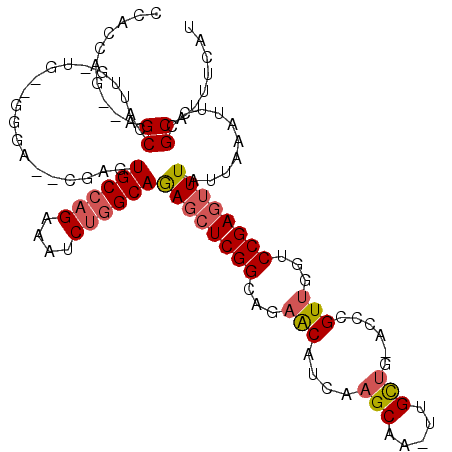
| Location | 19,236,010 – 19,236,122 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 97.02 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -35.77 |
| Energy contribution | -35.33 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19236010 112 + 20766785 CCCGUUGGUCCGAGUUUAUUAAAUUCCGCAUUUCAUUAUGAGUGCAUUGCCUCAUAGAGAGCUUCAUGCGGCUAUGUUGGCUUUGGGCUCAGACGCUCGGAUGGAAGUACUU ....((.((((((((.......................(((((.((..(((.(((((...((.....))..)))))..)))..)).)))))...)))))))).))....... ( -35.77) >DroSec_CAF1 26893 112 + 1 CCCGUUGGUCCGAGCUUAUUAAAUUCCGCAUUUCAUUAUGAGUGCAUUGCCUCAUAGAGAGCUCCAUGCGGCUAUGUUGGCUUUGGGCUCAGACGCUCGGAUGGAUGUACUC ..((((.((((((((.....((((.....)))).....(((((.((..(((.(((((...((.....))..)))))..)))..)).)))))...)))))))).))))..... ( -41.40) >DroSim_CAF1 25434 112 + 1 CCCGUUGGUCCGAGUUUAUUAAAUUCCGCAUUUCAUUAUGAGUGCAUUGCCUCAUAGAGAGCUCCAUGCGGCUAUGUUGGCUUUGGGCCCAGACGCUCGGAUGGAAGUACUU ....((.((((((((..........(((((((((.(((((((.((...))))))))).)))....))))))....((((((.....)))..))))))))))).))....... ( -35.70) >consensus CCCGUUGGUCCGAGUUUAUUAAAUUCCGCAUUUCAUUAUGAGUGCAUUGCCUCAUAGAGAGCUCCAUGCGGCUAUGUUGGCUUUGGGCUCAGACGCUCGGAUGGAAGUACUU ....((.((((((((..........(((((((((.(((((((.((...))))))))).)))....))))))....((((((.....)))..))))))))))).))....... (-35.77 = -35.33 + -0.44)


| Location | 19,236,046 – 19,236,150 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 96.79 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -29.08 |
| Energy contribution | -29.42 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19236046 104 - 20766785 ACUUUGGCUCUCAGCAACAGGCGUGCCAAAGUACUUCCAUCCGAGCGUCUGAGCCCAAAGCCAACAUAGCCGCAUGAAGCUCUCUAUGAGGCAAUGCACUCAUA ((((((((.....((.....))..)))))))).................((((..((..(((..(((((..((.....))...))))).)))..))..)))).. ( -26.10) >DroSec_CAF1 26929 104 - 1 ACUUUGGCUCUCAGCAACAGGCGUGCCCGAGUACAUCCAUCCGAGCGUCUGAGCCCAAAGCCAACAUAGCCGCAUGGAGCUCUCUAUGAGGCAAUGCACUCAUA ...((((((....((..(((((((...((..(......)..)).))))))).))....))))))(((.(((.((((((....)))))).))).)))........ ( -32.70) >DroSim_CAF1 25470 104 - 1 ACUUUGGCUCUCAGCAACAGGCGUGCCAAAGUACUUCCAUCCGAGCGUCUGGGCCCAAAGCCAACAUAGCCGCAUGGAGCUCUCUAUGAGGCAAUGCACUCAUA ((((((((.....((.....))..))))))))..........(((.((...(((.....)))......(((.((((((....)))))).)))...)).)))... ( -33.80) >consensus ACUUUGGCUCUCAGCAACAGGCGUGCCAAAGUACUUCCAUCCGAGCGUCUGAGCCCAAAGCCAACAUAGCCGCAUGGAGCUCUCUAUGAGGCAAUGCACUCAUA ...((((((....((..(((((((....................))))))).))....))))))(((.(((.((((((....)))))).))).)))........ (-29.08 = -29.42 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:07 2006