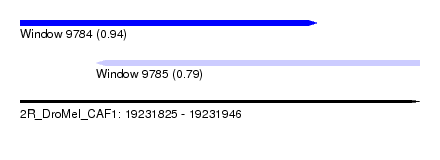
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,231,825 – 19,231,946 |
| Length | 121 |
| Max. P | 0.943500 |

| Location | 19,231,825 – 19,231,915 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -22.82 |
| Energy contribution | -22.77 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19231825 90 + 20766785 CCC----------------GAAUAUCAGAACAAACAGAUGUGCGCCAUUCGCACGCUGGCAGUAGUCCAAAAAGGCUUUCGCCUC-GUUCCGAGUGGCAAAGGGGCG (((----------------(.(((((..........))))).)((((((((.(((..(((...((((......))))...))).)-))..))))))))...)))... ( -31.40) >DroVir_CAF1 23791 94 + 1 -----------AUGAGCAACAAUAUUAA-AUAAACAGAUGUGCGCCAUUCGGAUUCUGGCAACAGUCCAAA-AGGCUUUUGCCUCUAGGGUGAGUGACAGAAUCACG -----------.................-.......(((.(((((((((((((..(((....))))))...-((((....))))...))))).))).))..)))... ( -24.00) >DroSec_CAF1 22952 90 + 1 GCC----------------GAAUAUCAGAACAAACAGAUGUGCGCCAUUCGCACGCUGGCAGGAGUCCAAAAAGGCUUUCGCCUC-GGGCCGAGUGGCAAAGGGGCA (((----------------(.(((((..........))))).)(((((((((.((..(((..(((((......)))))..))).)-).).)))))))).....))). ( -35.10) >DroSim_CAF1 21455 90 + 1 GUC----------------GAAUAUCAGAACAAACAGAUGUGCGCCAUUCGCACGCUGGCAGGAGUCCAAAAAGGCUUUCGCCUC-GGGCCGAGUGGCAAAGGGGCA (((----------------(.(((((..........))))).)(((((((((.((..(((..(((((......)))))..))).)-).).)))))))).....))). ( -32.40) >DroEre_CAF1 22566 86 + 1 -CC----------------AAAUAUCAGAGCAAACAGAUGUGCGCCAUUCGCACGCUGGCAGGAGUCCAAAAAGGCUUUCGCCUC-GGGCCGAGUGGCAAAGGC--- -((----------------..........(((........)))(((((((((.((..(((..(((((......)))))..))).)-).).))))))))...)).--- ( -30.90) >DroAna_CAF1 21261 102 + 1 GUCACCGUGGCAUGAG-AGCAAUAUCAAAAUAAACAGAUGUGCGCCAUUCGCACUCUGGCAGGAGUCCAAAAAGGCUUUCGCCUU-UGGCCGAGUGGUAAGCGU--- .........((.....-.(((.((((..........)))))))((((((((..(...(((..(((((......)))))..)))..-.)..))))))))..))..--- ( -31.40) >consensus GCC________________GAAUAUCAGAACAAACAGAUGUGCGCCAUUCGCACGCUGGCAGGAGUCCAAAAAGGCUUUCGCCUC_GGGCCGAGUGGCAAAGGGGC_ .....................(((((..........)))))..((((((((......(((.((((((......)))))).))).......))))))))......... (-22.82 = -22.77 + -0.05)

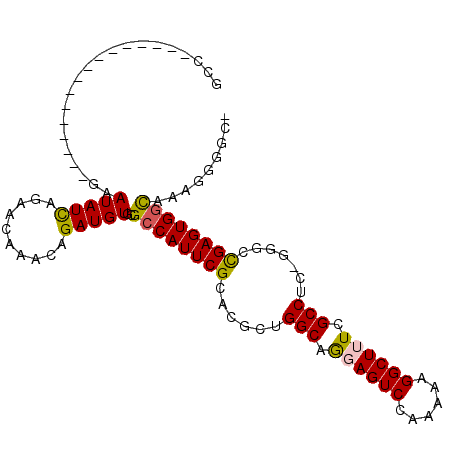
| Location | 19,231,848 – 19,231,946 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.91 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.02 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19231848 98 - 20766785 CCAGUCAAUCACUCAAUCAGCU-CAAUCA--------AGUCGCCCCUUUGCCACUCGGAAC-GAGGCGAAAGCCUUUUUGGACUACUGCCAGCGUGCGAAUGGCGCAC ...................((.-.....(--------((......))).((((.(((..((-(((((....))))..((((.(....)))))))).))).)))))).. ( -24.60) >DroVir_CAF1 23818 86 - 1 ---GUCAGUCA----------GUAUGUCA--------UGUCGUGAUUCUGUCACUCACCCUAGAGGCAAAAGCCU-UUUGGACUGUUGCCAGAAUCCGAAUGGCGCAC ---........----------((.(((((--------(.(((.(((((((.((..((.((.((((((....))))-)).))..)).)).)))))))))))))))).)) ( -29.20) >DroSec_CAF1 22975 106 - 1 CCAGUCAAUCACUCAAUCAGCC-CAAUCAGCUCAAUCAGCUGCCCCUUUGCCACUCGGCCC-GAGGCGAAAGCCUUUUUGGACUCCUGCCAGCGUGCGAAUGGCGCAC ...................(((-((..(((((.....)))))....((((((.((.(((((-(((((....)))))...))......))))).).)))))))).)).. ( -30.00) >DroSim_CAF1 21478 106 - 1 CCAGUCAAUCACUCAAUCAGCC-CAAUCAGCUCAAUCAGCUGCCCCUUUGCCACUCGGCCC-GAGGCGAAAGCCUUUUUGGACUCCUGCCAGCGUGCGAAUGGCGCAC ...................(((-((..(((((.....)))))....((((((.((.(((((-(((((....)))))...))......))))).).)))))))).)).. ( -30.00) >DroEre_CAF1 22588 86 - 1 CCAGUCAAUCACUCAAUCAGCC---------------------GCCUUUGCCACUCGGCCC-GAGGCGAAAGCCUUUUUGGACUCCUGCCAGCGUGCGAAUGGCGCAC ...................((.---------------------(((((((((.((.(((((-(((((....)))))...))......))))).).))))).))))).. ( -27.80) >DroYak_CAF1 22553 86 - 1 CCAGUCAAUCACUCAAUCAGCC---------------------GCCUUUGCCACUCGGAAC-GAGGCGAAAGCCUUUUUGGACUCCUGCCAGCGUGCGAAUGGCGCAC ...................((.---------------------((((((((.((.(.....-(((((....)))))..(((.(....))))).))))))).))))).. ( -25.20) >consensus CCAGUCAAUCACUCAAUCAGCC_CAAUCA________AG__GCCCCUUUGCCACUCGGCCC_GAGGCGAAAGCCUUUUUGGACUCCUGCCAGCGUGCGAAUGGCGCAC .................................................((((.(((.....(((((....))))).((((.(....)))))....))).)))).... (-19.82 = -20.02 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:01 2006