
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,228,504 – 19,228,622 |
| Length | 118 |
| Max. P | 0.858240 |

| Location | 19,228,504 – 19,228,622 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -39.85 |
| Consensus MFE | -23.35 |
| Energy contribution | -24.47 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19228504 118 - 20766785 UGGUCAUGGUAAACAU--GGCCGAGACAAUGUAGCUGCUUUUCGCUGGUUGGCCCAACUAAUUAGCGAAAGUAUUGAGCCAAGCCUCGGCCUGGUCCAGUCUACACCAGCACACUCCAGA .(((((((.....)))--))))(((....(((.((((...((((((((((((.....))))))))))))((.((((..(((.((....)).)))..))))))....)))))))))).... ( -45.30) >DroSec_CAF1 19608 118 - 1 UGGUCAUGGUAAACAU--GGCCGAGACAAUGUAGCUGCUUUUCGCUGGUUGGCCCAACUAAUUAGCGAAAGUAUUGAGCCAAGCCUCGGCCUGGUCCAGUCUACUCCAGCACACUCCAGA .(((((((.....)))--))))(((....(((.((((...((((((((((((.....))))))))))))((.((((..(((.((....)).)))..))))))....)))))))))).... ( -45.30) >DroSim_CAF1 18168 118 - 1 UGGUCAUGGUAAACAU--GGCCGAGACAAUGUAGCUGCUUUUCGCUGGUUGGCCCAACUAAUUAGCGAAAGUAUUGAGCCAAGCCUCGGCCUGGUCCAGUCUACUCCAGCGCACUCCAGA .(((((((.....)))--))))(((....(((.((((...((((((((((((.....))))))))))))((.((((..(((.((....)).)))..))))))....)))))))))).... ( -44.90) >DroEre_CAF1 19287 102 - 1 UGGUCAUGGUGAACAU--GGCCGAGACAAAGUAGCUGCUCUUCGCUGA-UGGCCCAACUAAUUAGCGAAAGUAUGGAGUCAAACCUCGGCCUGGUCC---------------ACUCCAGA .(((((((.....)))--)))).............((((.((((((((-(..........)))))))))))))((((((...(((.......)))..---------------)))))).. ( -34.10) >DroYak_CAF1 19036 118 - 1 UGGUCAUGGUAAACAU--GGCCGAGACAAAGUAGCUGCUUUUCGCUGGUUGGCCCAACUAAUUAGCGAAAGUAUUGAGCCGAACCCCGGCCUGGUCCAGUCUACUCCAGCACACUCCAGA .(((((((.....)))--))))(((....((((((((.((((((((((((((.....)))))))))))))).(((..((((.....))))..))).))).)))))........))).... ( -40.50) >DroAna_CAF1 17982 108 - 1 UC-----UGAAAACAAAGUGCCGAGU---UGUCGCUGGUAU----UGGUUUGCCCAACUGAUUAGCAACGAUAUUGGACUAAACUUCGGCCUACUCCGGCCCAUACCAGCCCAGACCAGA ((-----((.......(((.((((((---(((.((((((.(----(((.....))))...)))))).))))).))))))).......((((......))))..........))))..... ( -29.00) >consensus UGGUCAUGGUAAACAU__GGCCGAGACAAUGUAGCUGCUUUUCGCUGGUUGGCCCAACUAAUUAGCGAAAGUAUUGAGCCAAACCUCGGCCUGGUCCAGUCUACUCCAGCACACUCCAGA ..................(((((((..........((((.((((((((((((.....))))))))))))))))...........)))))))............................. (-23.35 = -24.47 + 1.11)

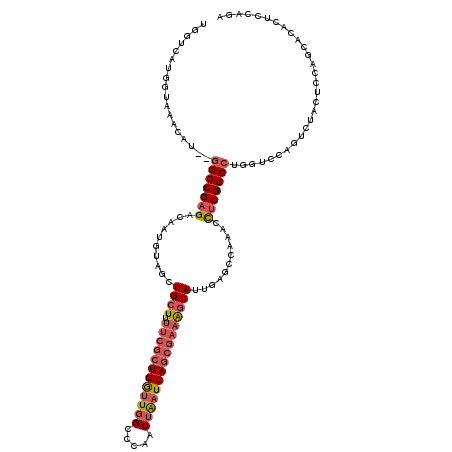
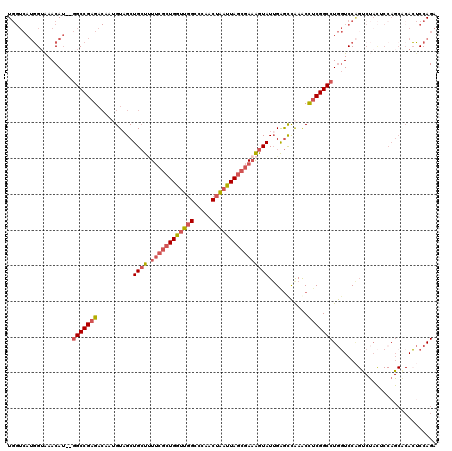
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:59 2006