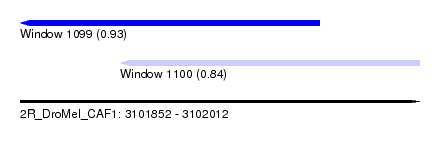
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,101,852 – 3,102,012 |
| Length | 160 |
| Max. P | 0.931517 |

| Location | 3,101,852 – 3,101,972 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.89 |
| Mean single sequence MFE | -40.65 |
| Consensus MFE | -30.27 |
| Energy contribution | -31.95 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3101852 120 - 20766785 AGUUGCCAAGGGCUGCCACUUUGUCGUAUUUGGCAGGGUGGCAGCUCUGUCGCACAUCAAUGGUUAAACCGGUUCACCAAUCACACAUAUUUACCGGUUUAAUUUCGGCCGACAGCUGUU .((((((.((((((((((((((((((....)))))))))))))))))).............((((((((((((...................))))))))))))..)).))))....... ( -50.81) >DroSec_CAF1 16489 120 - 1 AGUUGCCAGUGGCUGCCACACUGUCGGAUUUCGCAGGGUGGCAGCUCUGUUUCACAUCAAUGGUUGAACCGGUUCGGCAAUCGCACAUAUUUACCGGUUUAAUUUCGGCCGACAGCUGUU (((((.(((.(((((((((.(((.((.....))))).))))))))))))...........(((((((((((((...((....))........))))))......))))))).)))))... ( -44.10) >DroSim_CAF1 23209 120 - 1 AGUUGCCAGUGGGUGCCACACUGUCGUAUUUAGCAGGGUGGCAGCUCUGUCUCACAUCAAUGGUUGAACCGGUUCGGCAAUCGCACAUAUUUACCGGUUUAAUUUCGGCCGACAGCUGUU .((((((.(((((((((((.((((........)))).)))))).......)))))......((((((((((((...((....))........))))))))))))..)).))))....... ( -40.31) >DroEre_CAF1 16669 117 - 1 AGUUUGCAGUGGCUGCCACACUAUCGUAUUUCGCAGGGUGGCAGCUCUGU--CACAUCGGUUUUUAAACCGGUUCACUAAUCGCACAUAUUUUCCGGUUACA-GUUGCCCGACGGCUGUU .((..((((.(((((((((.((..((.....))..)))))))))))))))--.))(((((((....)))))))....((((((...........))))))((-((((.....)))))).. ( -36.10) >DroYak_CAF1 16706 118 - 1 AGUUUCCAUUAGCUGCCACCAUAUCGGAUUUCGCAGGGUGGCAGCUCUGU--CACAUCGAUUAUUAAACCGGUUCGCCAAUCGCACAUAUUUACCGGAUAAAUGUAGACUGCCAGCUGUU (((((.((((((((((((((....((.....))...))))))))))....--................(((((...................)))))...)))).))))).......... ( -31.91) >consensus AGUUGCCAGUGGCUGCCACACUGUCGUAUUUCGCAGGGUGGCAGCUCUGU__CACAUCAAUGGUUAAACCGGUUCGCCAAUCGCACAUAUUUACCGGUUUAAUUUCGGCCGACAGCUGUU (((((.(((.(((((((((.((((........)))).))))))))))))............((((((((((((...................))))))))))))........)))))... (-30.27 = -31.95 + 1.68)

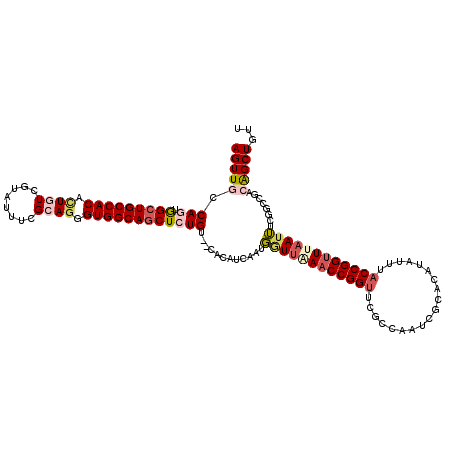
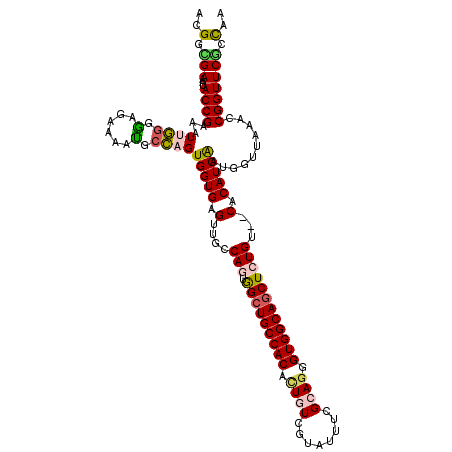
| Location | 3,101,892 – 3,102,012 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -42.24 |
| Consensus MFE | -28.66 |
| Energy contribution | -28.94 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3101892 120 - 20766785 ACGACGAAAUACCGAAAUUGGGGAGAAAAUGCCAGUGGUGAGUUGCCAAGGGCUGCCACUUUGUCGUAUUUGGCAGGGUGGCAGCUCUGUCGCACAUCAAUGGUUAAACCGGUUCACCAA ....((......))...((((.(((.....((((.(((((...(((..((((((((((((((((((....))))))))))))))))))...)))))))).))))........))).)))) ( -48.62) >DroSec_CAF1 16529 120 - 1 ACGGCGAAAAACCGAAAUUGGGGAGAAAAUGCCAGUGGUGAGUUGCCAGUGGCUGCCACACUGUCGGAUUUCGCAGGGUGGCAGCUCUGUUUCACAUCAAUGGUUGAACCGGUUCGGCAA ..(.(((...((((((((.(((........((((.((((.....)))).))))((((((.(((.((.....))))).)))))).))).)))))........((.....)))))))).).. ( -44.20) >DroSim_CAF1 23249 120 - 1 ACGGCGAAAAACCGAAAUUGGGGAGAAAAUGCCAGUGGUGAGUUGCCAGUGGGUGCCACACUGUCGUAUUUAGCAGGGUGGCAGCUCUGUCUCACAUCAAUGGUUGAACCGGUUCGGCAA ..(.(((...(((...(((((.((((.....(((.((((.....)))).))).((((((.((((........)))).))))))......))))...)))))((.....)))))))).).. ( -41.40) >DroEre_CAF1 16708 118 - 1 ACGAUGAAAUACCGAAUUUGUGGAGAACACGCCUGUGGUGAGUUUGCAGUGGCUGCCACACUAUCGUAUUUCGCAGGGUGGCAGCUCUGU--CACAUCGGUUUUUAAACCGGUUCACUAA .((...(((.(((((...((((..((((.((((...)))).))))((((.(((((((((.((..((.....))..)))))))))))))))--))))))))).)))....))......... ( -39.80) >DroYak_CAF1 16746 118 - 1 ACGGUGAAAUACCGAAUUUAGGUAGAACAAGCUCGUGGUGAGUUUCCAUUAGCUGCCACCAUAUCGGAUUUCGCAGGGUGGCAGCUCUGU--CACAUCGAUUAUUAAACCGGUUCGCCAA ..((((((.((((.......))))........(((..((((.....((..((((((((((....((.....))...)))))))))).)))--)))..)))............)))))).. ( -37.20) >consensus ACGGCGAAAUACCGAAAUUGGGGAGAAAAUGCCAGUGGUGAGUUGCCAGUGGCUGCCACACUGUCGUAUUUCGCAGGGUGGCAGCUCUGU__CACAUCAAUGGUUAAACCGGUUCGCCAA ..(.(((...((((...((((.(......).))))(((((.(....(((.(((((((((.((((........)))).))))))))))))...).)))))..........))))))).).. (-28.66 = -28.94 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:47 2006