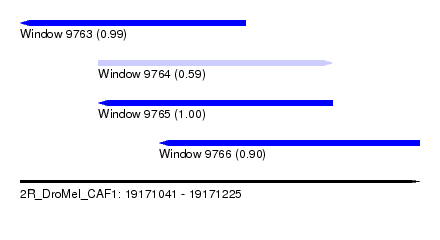
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,171,041 – 19,171,225 |
| Length | 184 |
| Max. P | 0.999895 |

| Location | 19,171,041 – 19,171,145 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -34.31 |
| Consensus MFE | -28.18 |
| Energy contribution | -29.25 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19171041 104 - 20766785 AGUGUGACCUUGCUCUUAAAGUAAGGUCCAAAUUAUUUUGCGAGCUAAGGUCACGCUUAUUUUGAGCU----------------AAGUUCCUCCCACAUUUUUCUCCGUGCAGCAUUCGA (((((((((((((((.((((((((........)))))))).)))).)))))))))))......(((..----------------((((.........))))..))).............. ( -29.20) >DroSec_CAF1 54848 104 - 1 AGUGUGACCUUGCUCUCAAAGUAAGGUCCAAAUUAGUUUGCCAGCUAAGGUCACGCCUAUCUUGAGCU----------------AAGUUGCUCCUACAUUUUUCUCCGUGCAGCAUCCGA .(((((((((((((..((((.(((........))).))))..))).))))))))))............----------------..(((((..................)))))...... ( -29.37) >DroEre_CAF1 62423 120 - 1 AGUGUGACCUUGCUCUUAAAGUAGGGUCCAAAUUAUUUCGCGAGCUAAGGUCACGCUUAGCUUGAGCUAAGGCACUCGCCUUUUAAGUUGCUUGUACAUUUUUCUCCGUGCAGCAUUCGA (((((((((((((((...((((((........))))))...)))).))))))))))).(((((((...(((((....))))))))))))(((.((((..........)))))))...... ( -40.80) >DroYak_CAF1 61917 120 - 1 AGUGUGACCUUGCUCUCAAAGUAAGGUCCAAAUUAUUUUGCGUGCUAUGGUCACGCUUAGCUUGAGCUGAGGCAUUUGCCUUCAAAGUUGCUCCUACUUUUUUCUCCGUGCAGCAUCCGA .((((((((..((.(.((((((((........)))))))).).))...))))))))(((((....)))))(((....)))......(((((..................)))))...... ( -37.87) >consensus AGUGUGACCUUGCUCUCAAAGUAAGGUCCAAAUUAUUUUGCGAGCUAAGGUCACGCUUAGCUUGAGCU________________AAGUUGCUCCUACAUUUUUCUCCGUGCAGCAUCCGA .((((((((((((((.((((((((........)))))))).)))).))))))))))..............................(((((..................)))))...... (-28.18 = -29.25 + 1.06)

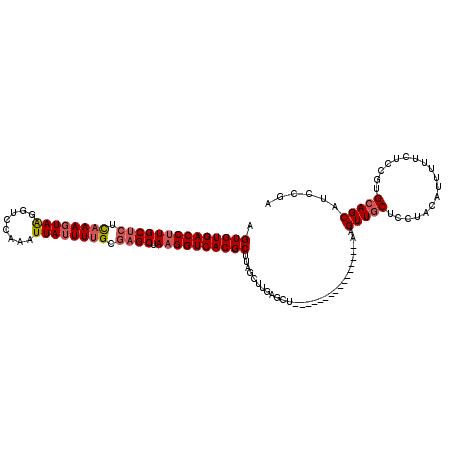
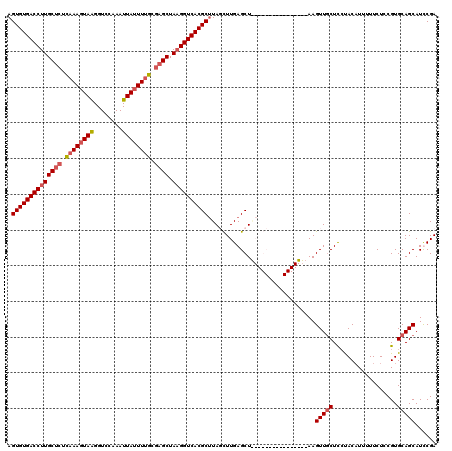
| Location | 19,171,077 – 19,171,185 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -21.65 |
| Energy contribution | -24.15 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19171077 108 + 20766785 ------------AGCUCAAAAUAAGCGUGACCUUAGCUCGCAAAAUAAUUUGGACCUUACUUUAAGAGCAAGGUCACACUGCAUAUAGAUUAAAGUGCCUGUGCAAAGGAUUGGAGUUUG ------------(((((......((.((((((((.((((..(((.(((........))).)))..)))))))))))).))(((((..............))))).........))))).. ( -27.74) >DroSec_CAF1 54884 95 + 1 ------------AGCUCAAGAUAGGCGUGACCUUAGCUGGCAAACUAAUUUGGACCUUACUUUGAGAGCAAGGUCACACUGCACCUAGAUUAAAGGGCCUG-------------AGUUUG ------------((((((.....(((((((((((.(((..((((.(((........))).))))..)))))))))))......(((.......))))))))-------------)))).. ( -30.40) >DroEre_CAF1 62463 120 + 1 GGCGAGUGCCUUAGCUCAAGCUAAGCGUGACCUUAGCUCGCGAAAUAAUUUGGACCCUACUUUAAGAGCAAGGUCACACUGCAGCUAGCCUAAAGGGCCUAUGCAAGGGAUUGGAGUUUG ..(.(((.((((.((...((((.((.((((((((.((((..(((.((..........)).)))..)))))))))))).))..)))).(((.....)))....)))))).))).)...... ( -38.50) >DroYak_CAF1 61957 120 + 1 GGCAAAUGCCUCAGCUCAAGCUAAGCGUGACCAUAGCACGCAAAAUAAUUUGGACCUUACUUUGAGAGCAAGGUCACACUGCAGCCAGCUUAAAGGACCUAAGCAAAGGAUUGGAGUUUG (((....)))..(((((..(((.((.((((((...((.(.((((.(((........))).)))).).))..)))))).))..)))..(((((.......))))).........))))).. ( -31.90) >consensus ____________AGCUCAAGAUAAGCGUGACCUUAGCUCGCAAAAUAAUUUGGACCUUACUUUAAGAGCAAGGUCACACUGCAGCUAGAUUAAAGGGCCUAUGCAAAGGAUUGGAGUUUG ............(((((.((((.((.((((((((.((((.((((.(((........))).)))).)))))))))))).))..))))...........(((......)))....))))).. (-21.65 = -24.15 + 2.50)

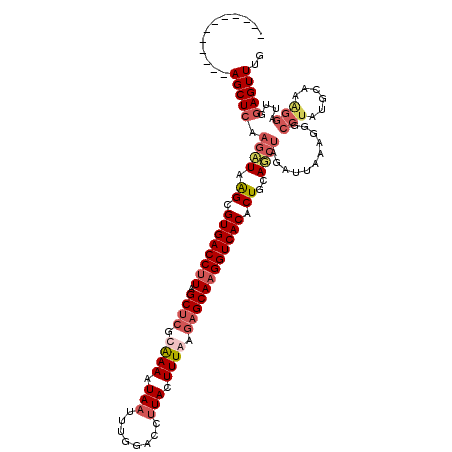
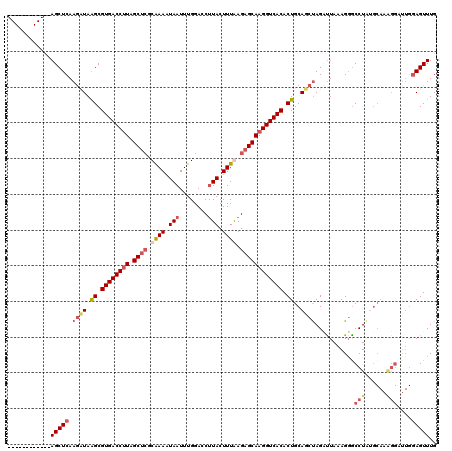
| Location | 19,171,077 – 19,171,185 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -29.35 |
| Energy contribution | -30.23 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.79 |
| SVM decision value | 4.42 |
| SVM RNA-class probability | 0.999895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19171077 108 - 20766785 CAAACUCCAAUCCUUUGCACAGGCACUUUAAUCUAUAUGCAGUGUGACCUUGCUCUUAAAGUAAGGUCCAAAUUAUUUUGCGAGCUAAGGUCACGCUUAUUUUGAGCU------------ ................((.((((((..(......)..)))(((((((((((((((.((((((((........)))))))).)))).)))))))))))....))).)).------------ ( -32.70) >DroSec_CAF1 54884 95 - 1 CAAACU-------------CAGGCCCUUUAAUCUAGGUGCAGUGUGACCUUGCUCUCAAAGUAAGGUCCAAAUUAGUUUGCCAGCUAAGGUCACGCCUAUCUUGAGCU------------ ....((-------------((((((((.......))).)).(((((((((((((..((((.(((........))).))))..))).)))))))))).....)))))..------------ ( -32.80) >DroEre_CAF1 62463 120 - 1 CAAACUCCAAUCCCUUGCAUAGGCCCUUUAGGCUAGCUGCAGUGUGACCUUGCUCUUAAAGUAGGGUCCAAAUUAUUUCGCGAGCUAAGGUCACGCUUAGCUUGAGCUAAGGCACUCGCC ............((((((...((((.....))))(((((.(((((((((((((((...((((((........))))))...)))).))))))))))))))))...)).))))........ ( -41.40) >DroYak_CAF1 61957 120 - 1 CAAACUCCAAUCCUUUGCUUAGGUCCUUUAAGCUGGCUGCAGUGUGACCUUGCUCUCAAAGUAAGGUCCAAAUUAUUUUGCGUGCUAUGGUCACGCUUAGCUUGAGCUGAGGCAUUUGCC .......(((.((((.((((((((....((((((((((..((..((((((((((.....)))))))))((((....)))).)..))..))))).))))))))))))).))))...))).. ( -41.50) >consensus CAAACUCCAAUCCUUUGCACAGGCCCUUUAAGCUAGCUGCAGUGUGACCUUGCUCUCAAAGUAAGGUCCAAAUUAUUUUGCGAGCUAAGGUCACGCUUAGCUUGAGCU____________ ..............................(((((((((.(((((((((((((((.((((((((........)))))))).)))).))))))))))))))))..))))............ (-29.35 = -30.23 + 0.87)


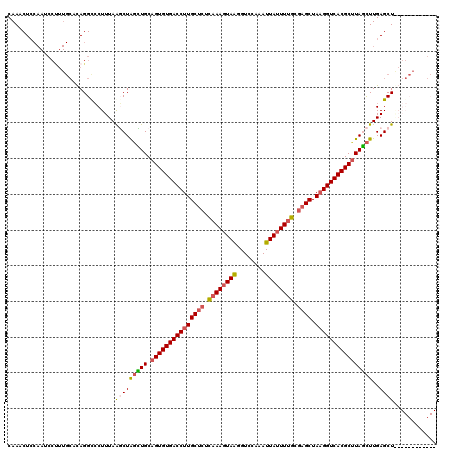
| Location | 19,171,105 – 19,171,225 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.30 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -16.58 |
| Energy contribution | -16.77 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19171105 120 - 20766785 AGUUGAAUUUGAAAUGAUUAAAGGCUAACUUUUGCCAUGCCAAACUCCAAUCCUUUGCACAGGCACUUUAAUCUAUAUGCAGUGUGACCUUGCUCUUAAAGUAAGGUCCAAAUUAUUUUG .....((((((....((((((((((........))).((((......(((....)))....)))).)))))))............(((((((((.....)))))))))))))))...... ( -28.80) >DroSec_CAF1 54912 100 - 1 CUUUGCAUUUUAAAUGAUUAAAUGAU-------UUCAUGCCAAACU-------------CAGGCCCUUUAAUCUAGGUGCAGUGUGACCUUGCUCUCAAAGUAAGGUCCAAAUUAGUUUG ..((((((((.....(((((((((..-------..)))(((.....-------------..)))...)))))).))))))))((.(((((((((.....))))))))))).......... ( -26.70) >DroEre_CAF1 62503 118 - 1 CUUUGAAUUUUAAAUGAUUC-AG-CUUUUUUUAUCCAUGCCAAACUCCAAUCCCUUGCAUAGGCCCUUUAGGCUAGCUGCAGUGUGACCUUGCUCUUAAAGUAGGGUCCAAAUUAUUUCG ...........(((((((((-((-((..........((((................)))).((((.....)))))))))...((.(((((((((.....))))))))))))))))))).. ( -26.19) >DroYak_CAF1 61997 119 - 1 CUUUGAAUUUUAAAUGCUUAAAG-CUUUUUUCGUCCAAGCCAAACUCCAAUCCUUUGCUUAGGUCCUUUAAGCUGGCUGCAGUGUGACCUUGCUCUCAAAGUAAGGUCCAAAUUAUUUUG ..............(((....((-(((.....(.((((((.(((.........))))))).)).)....)))))....))).((.(((((((((.....))))))))))).......... ( -26.30) >consensus CUUUGAAUUUUAAAUGAUUAAAG_CUUUUUUU_UCCAUGCCAAACUCCAAUCCUUUGCACAGGCCCUUUAAGCUAGCUGCAGUGUGACCUUGCUCUCAAAGUAAGGUCCAAAUUAUUUUG ...............((((((((...............(((....................))).)))))))).........((.(((((((((.....))))))))))).......... (-16.58 = -16.77 + 0.19)

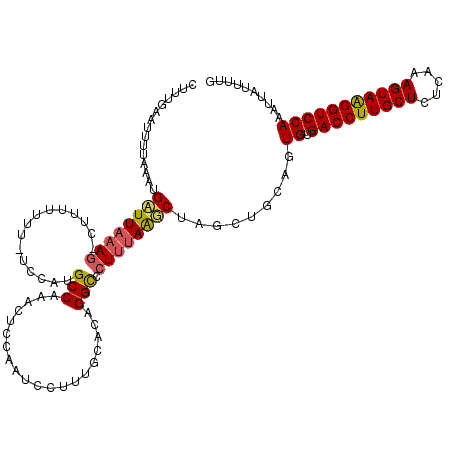

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:43 2006