| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,168,597 – 19,168,695 |
| Length | 98 |
| Max. P | 0.527043 |

| Location | 19,168,597 – 19,168,695 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.26 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -8.93 |
| Energy contribution | -9.98 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.39 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19168597 98 - 20766785 G----CAGCGCAA----UCC-GAAAUGCGAAA-------UGCAAAAUGCAAAACACGAAUGGGGCGAAAUGCGAACGCAGUUUGCAAACUUAUCGCC---UCCUGUUGUUUAUAAGU (----((.((((.----...-....))))...-------)))........(((((((...(((((((..(((((((...)))))))......)))))---)).)).)))))...... ( -26.60) >DroSec_CAF1 52475 86 - 1 G----CGGCGCAA------------UGCGAAA-------UGCAAAAUGCAAAACACGAAUG--GCGAAAUGCGAACGCAGUUUGCAAACUUAUCGCC---GCCUG---UUUAUAAGU (----(((((((.------------(((....-------.)))...)))............--(((((.(((....))).))))).........)))---))...---......... ( -24.00) >DroEre_CAF1 60025 96 - 1 G----CAGCGAAA----UGC-GAAAUGCGAAA-------UGCAAAAUGCAAAACACGAAUG--GCGAAAUGCGAACGCAGUUUGCAAACUUAUCGCCGCCGCCAG---UUUAUAAGU (----(.((....----(((-(...(((....-------.)))...))))..........(--((((..(((((((...)))))))......))))))).))...---......... ( -22.80) >DroYak_CAF1 59445 100 - 1 G----CAGUGAAA----UGC-AAAAUGCGAAAUGCUGAAUGCAAAAUGCAAAACACGAAUG--GCGAAAUGCGAACGCAGUUUGCAAACUUAUCGCC---GUCUG---UUUAUAAGU (----((......----)))-....((((...(((.....)))...))))(((((.((.((--((((..(((((((...)))))))......)))))---)))))---)))...... ( -26.60) >DroAna_CAF1 62574 98 - 1 GCAGUCAGGGAGA----AGCUAAAAUGCAAAA-------UGCAAAAUUCGAAACACGAAAA--ACGAAAUGCGAACGCAAUUUGCAAACUUAUUGUU---GCUUG---UUUAUAAGU ((((.(((.(((.----.((......))....-------((((((.((((.....))))..--......(((....))).))))))..))).)))))---))...---......... ( -19.60) >DroPer_CAF1 63262 86 - 1 A----AAUCGAAACGAGUCU-GAAAU----AA-------UGAAAAAUGAAAAAU---------GCAAAAUGCGAACGCCAAUUGCAAAUUUAUUGUG---GCUUG---UUUAUAAGU .----.....(((((((((.-...((----((-------((((...(....).(---------((((...((....))...)))))..)))))))))---)))))---)))...... ( -17.00) >consensus G____CAGCGAAA____UGC_GAAAUGCGAAA_______UGCAAAAUGCAAAACACGAAUG__GCGAAAUGCGAACGCAGUUUGCAAACUUAUCGCC___GCCUG___UUUAUAAGU ..........................((((.........(((.....))).............(((((.(((....))).))))).......))))..................... ( -8.93 = -9.98 + 1.06)

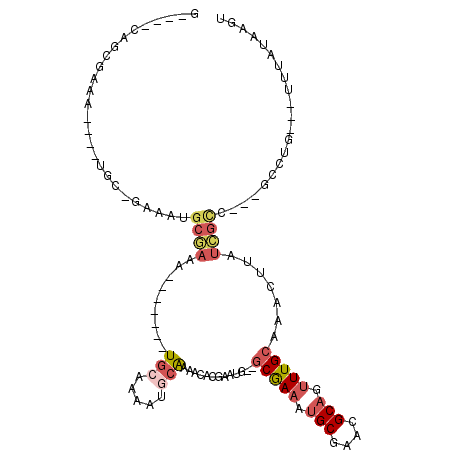
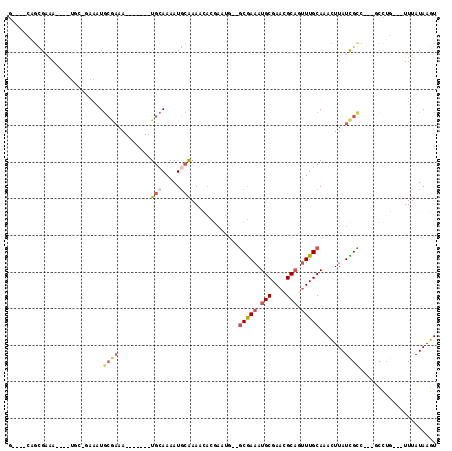
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:37 2006