| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,160,622 – 19,160,712 |
| Length | 90 |
| Max. P | 0.704406 |

| Location | 19,160,622 – 19,160,712 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 80.09 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -16.23 |
| Energy contribution | -15.40 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
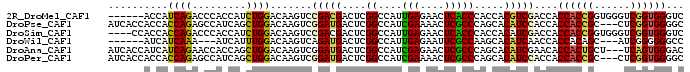
>2R_DroMel_CAF1 19160622 90 + 20766785 ------ACCAUCAGACCCACCAUCUGGACAAGUCCGACGACUCGGCCAUUGAGAACUCACCCACCACGUCGACCACCACCGGUGGGUCGGUGGGUC ------.......((((((((....(((....)))((((((..((....(((....)))....))..)))..(((((...)))))))))))))))) ( -35.70) >DroPse_CAF1 54852 93 + 1 AUCACCACCACCAGAGCCAUCAGCUGGACAAGUCGGAUGACUCGGCCAUCGAAAACUCGCCCAGCACAUCCACCACCACCGC---CUCGGUGGGGC ...............(((....(((((....(((((.....)))))...(((....))).)))))..........((((((.---..))))))))) ( -27.00) >DroSim_CAF1 45418 92 + 1 ----CCACCACCAGACCCACCAUCUGGACAAGUCCGACGACUCGGCCAUUGAGAACUCACCCACCACAUCGACCACCACCGGUGGGUCGGUGGGUC ----...(((((.((((((((...((((....)))).(((...((....(((....)))....))...))).........)))))))))))))... ( -35.00) >DroWil_CAF1 70369 84 + 1 ------AUCAUCAAA---AUCAUUUGGACAAGUCAGAUGACUCGGCCAUUGAGAAUUCGCCAAGCACAUCAACCACCACAGC---AUCGGGGGGCC ------.........---.((((((((.....))))))))...((((.((((......((...))...))))((.((.....---...)))))))) ( -17.20) >DroAna_CAF1 53575 93 + 1 AUCACCAUCAUCAGAACCACCAGCUGGACAAGUCGGAUGACUCGGCCAUCGAGAACUCGCCCAGCACAUCGAACACCACUGCU---UCAGUGGGAC ......................(((((.(.((((.((((.......))))..).))).).)))))..........((((((..---.))))))... ( -22.50) >DroPer_CAF1 54867 93 + 1 AUCACCACCACCAGAGCCAUCAGCUGGACAAGUCGGAUGACUCGGCCAUCGAAAACUCGCCCAGCACAUCCACCACCACCGC---CUCGGUGGGGC ...............(((....(((((....(((((.....)))))...(((....))).)))))..........((((((.---..))))))))) ( -27.00) >consensus ____CCACCACCAGACCCACCAGCUGGACAAGUCGGAUGACUCGGCCAUCGAGAACUCGCCCAGCACAUCGACCACCACCGC___CUCGGUGGGGC ..........((((.........)))).......(((((....((....(((....))))).....)))))....((((((......))))))... (-16.23 = -15.40 + -0.83)
| Location | 19,160,622 – 19,160,712 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 80.09 |
| Mean single sequence MFE | -38.35 |
| Consensus MFE | -21.87 |
| Energy contribution | -21.57 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
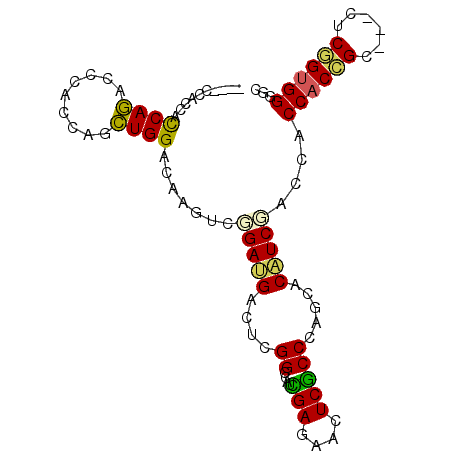
>2R_DroMel_CAF1 19160622 90 - 20766785 GACCCACCGACCCACCGGUGGUGGUCGACGUGGUGGGUGAGUUCUCAAUGGCCGAGUCGUCGGACUUGUCCAGAUGGUGGGUCUGAUGGU------ (((((((((((((((((((.(....).)).))))))))..........(((.((((((....)))))).)))..))))))))).......------ ( -44.20) >DroPse_CAF1 54852 93 - 1 GCCCCACCGAG---GCGGUGGUGGUGGAUGUGCUGGGCGAGUUUUCGAUGGCCGAGUCAUCCGACUUGUCCAGCUGAUGGCUCUGGUGGUGGUGAU (((((((((..---((.((.(..........((((((((((((...((((((...)))))).))))))))))))).)).))..)))))).)))... ( -43.10) >DroSim_CAF1 45418 92 - 1 GACCCACCGACCCACCGGUGGUGGUCGAUGUGGUGGGUGAGUUCUCAAUGGCCGAGUCGUCGGACUUGUCCAGAUGGUGGGUCUGGUGGUGG---- ..(((((((((((((((.(((.((((((((((((.(.(((....))).).))))...)))).))))...)))..))))))))).))))).).---- ( -44.90) >DroWil_CAF1 70369 84 - 1 GGCCCCCCGAU---GCUGUGGUGGUUGAUGUGCUUGGCGAAUUCUCAAUGGCCGAGUCAUCUGACUUGUCCAAAUGAU---UUUGAUGAU------ .......(((.---.(..(((.((((((((.(((((((.(........).))))))))))).))))...)))...)..---.))).....------ ( -21.30) >DroAna_CAF1 53575 93 - 1 GUCCCACUGA---AGCAGUGGUGUUCGAUGUGCUGGGCGAGUUCUCGAUGGCCGAGUCAUCCGACUUGUCCAGCUGGUGGUUCUGAUGAUGGUGAU (((((((((.---..)))))).(..(.((..((((((((((((...((((((...)))))).))))))))))))..)).)..).)))......... ( -33.50) >DroPer_CAF1 54867 93 - 1 GCCCCACCGAG---GCGGUGGUGGUGGAUGUGCUGGGCGAGUUUUCGAUGGCCGAGUCAUCCGACUUGUCCAGCUGAUGGCUCUGGUGGUGGUGAU (((((((((..---((.((.(..........((((((((((((...((((((...)))))).))))))))))))).)).))..)))))).)))... ( -43.10) >consensus GACCCACCGAC___GCGGUGGUGGUCGAUGUGCUGGGCGAGUUCUCAAUGGCCGAGUCAUCCGACUUGUCCAGAUGAUGGCUCUGAUGGUGG____ ..(((((((......)))))).).((((((.(((.(((.(........).))).))))))))).......(((.........)))........... (-21.87 = -21.57 + -0.30)

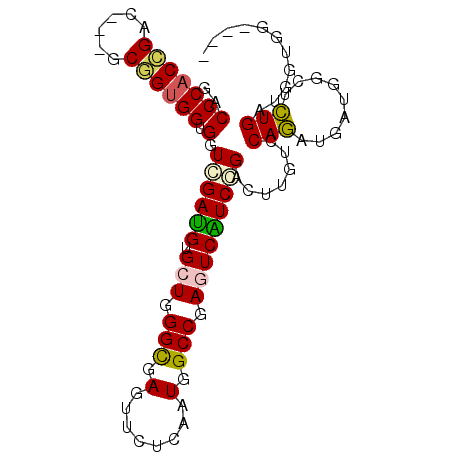
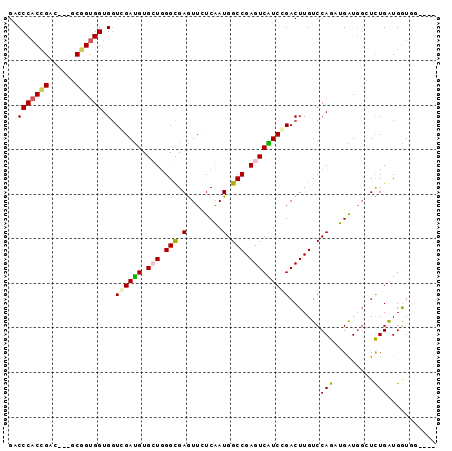
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:34 2006