| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,148,206 – 19,148,309 |
| Length | 103 |
| Max. P | 0.898212 |

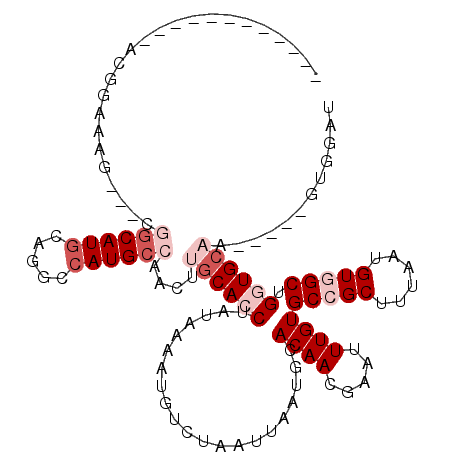
| Location | 19,148,206 – 19,148,309 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -20.89 |
| Energy contribution | -22.12 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19148206 103 + 20766785 AUCCAC-----UUGCACCAGCCACAUUAAAGCGGCACAAAUUCGUUGUGCAUUAAUUAGACAUUUUAUAGGUGCAAGUUGGCAUGGCCUGCAUGCCGCCGCUUUCCGU------------ ....((-----(((((((.((.........)).((((((.....))))))...................))))))))).((((((.....))))))............------------ ( -32.00) >DroSec_CAF1 32048 103 + 1 AUCCAC-----UUGCACCAGCCACAUUAAAGCGGCACAAAUUCGUUGUGCAUUAAUUAGACAUUUUAUAGGUGCAAGUUGGCAUGGCCUGCAUGCCGCCGCUUUCCGC------------ ....((-----(((((((.((.........)).((((((.....))))))...................))))))))).((((((.....))))))............------------ ( -32.00) >DroEre_CAF1 40209 112 + 1 AUCCAC-----UUGCACCAGCCACAUUAAAGCGGCACAAAUUCGUUGUGCAUUAAUUAGACAUUUUAUAGGUGCAAGUUGGCAUGGCCUGUAUGCCG---CUUUCGGUUGCCAGUUGCCA ....((-----(((((((.((.........)).((((((.....))))))...................)))))))))(((((((((......((((---....)))).))))..))))) ( -36.50) >DroYak_CAF1 38731 110 + 1 AUCCAC-----UUGCAACCGCCACAUUAAAGCGGCACAAAUUCGUUGUGCAUUAAUUAGACAUUUUAUAGGUGCAAGUUGGCAUGGCCUGCAUGCCG---CUUUUCGUUGGCCGUUGC-- ......-----..(((((.((((....(((((((((.......((..(((..(((((.(.((((.....))))).))))))))..)).....)))))---))))....)))).)))))-- ( -36.70) >DroAna_CAF1 41323 82 + 1 A-------------CACCAGCAACAUUAAAGCGGCACAAAUUCGUUGUGCAUUAAUUAGACAUUUUAUAGGUGCAAGUUGGCAUGGCCUGCAUGC------------------------- .-------------((((.((.........)).((((((.....))))))...................)))).......(((((.....)))))------------------------- ( -17.80) >DroPer_CAF1 41290 102 + 1 AACGACUGCAGAGGCAGCAGCAACAA-AAAGCGGCACAAAUUCGUUGUGCAUUAAUUAGACAUUUUAUAGGUGCAAGUUGGCAUGGCCUGCAUGUACCUG------GU-G---------- ...(.((((....))))).((.....-...)).((((((.....)))))).................((((((((.((.(((...))).)).))))))))------..-.---------- ( -29.40) >consensus AUCCAC_____UUGCACCAGCCACAUUAAAGCGGCACAAAUUCGUUGUGCAUUAAUUAGACAUUUUAUAGGUGCAAGUUGGCAUGGCCUGCAUGCCG___CUUUCCGU____________ ...........(((((((.((.........)).((((((.....))))))...................)))))))...((((((.....))))))........................ (-20.89 = -22.12 + 1.22)

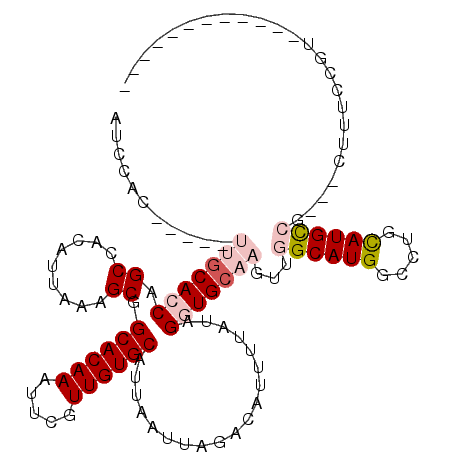
| Location | 19,148,206 – 19,148,309 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -18.28 |
| Energy contribution | -20.12 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19148206 103 - 20766785 ------------ACGGAAAGCGGCGGCAUGCAGGCCAUGCCAACUUGCACCUAUAAAAUGUCUAAUUAAUGCACAACGAAUUUGUGCCGCUUUAAUGUGGCUGGUGCAA-----GUGGAU ------------............((((((.....)))))).(((((((((.....................((((.....))))(((((......))))).)))))))-----)).... ( -33.20) >DroSec_CAF1 32048 103 - 1 ------------GCGGAAAGCGGCGGCAUGCAGGCCAUGCCAACUUGCACCUAUAAAAUGUCUAAUUAAUGCACAACGAAUUUGUGCCGCUUUAAUGUGGCUGGUGCAA-----GUGGAU ------------((.(....).))((((((.....)))))).(((((((((.....................((((.....))))(((((......))))).)))))))-----)).... ( -35.40) >DroEre_CAF1 40209 112 - 1 UGGCAACUGGCAACCGAAAG---CGGCAUACAGGCCAUGCCAACUUGCACCUAUAAAAUGUCUAAUUAAUGCACAACGAAUUUGUGCCGCUUUAAUGUGGCUGGUGCAA-----GUGGAU (((((..((((..(((....---))).......)))))))))(((((((((.....................((((.....))))(((((......))))).)))))))-----)).... ( -37.90) >DroYak_CAF1 38731 110 - 1 --GCAACGGCCAACGAAAAG---CGGCAUGCAGGCCAUGCCAACUUGCACCUAUAAAAUGUCUAAUUAAUGCACAACGAAUUUGUGCCGCUUUAAUGUGGCGGUUGCAA-----GUGGAU --(((((.((((....((((---(((((((((((.........)))))..........((((........).)))........))))))))))....)))).)))))..-----...... ( -38.10) >DroAna_CAF1 41323 82 - 1 -------------------------GCAUGCAGGCCAUGCCAACUUGCACCUAUAAAAUGUCUAAUUAAUGCACAACGAAUUUGUGCCGCUUUAAUGUUGCUGGUG-------------U -------------------------(((((.....)))))......(((((...................((((((.....)))))).((.........)).))))-------------) ( -17.60) >DroPer_CAF1 41290 102 - 1 ----------C-AC------CAGGUACAUGCAGGCCAUGCCAACUUGCACCUAUAAAAUGUCUAAUUAAUGCACAACGAAUUUGUGCCGCUUU-UUGUUGCUGCUGCCUCUGCAGUCGUU ----------.-..------.((((.((.(..(((...)))..).)).))))..................((((((.....))))))......-.....((.(((((....))))).)). ( -20.50) >consensus ____________ACGGAAAG___CGGCAUGCAGGCCAUGCCAACUUGCACCUAUAAAAUGUCUAAUUAAUGCACAACGAAUUUGUGCCGCUUUAAUGUGGCUGGUGCAA_____GUGGAU ........................((((((.....))))))....((((((.....................((((.....))))(((((......))))).))))))............ (-18.28 = -20.12 + 1.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:24 2006