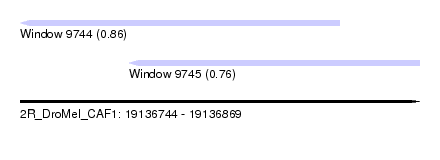
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,136,744 – 19,136,869 |
| Length | 125 |
| Max. P | 0.859150 |

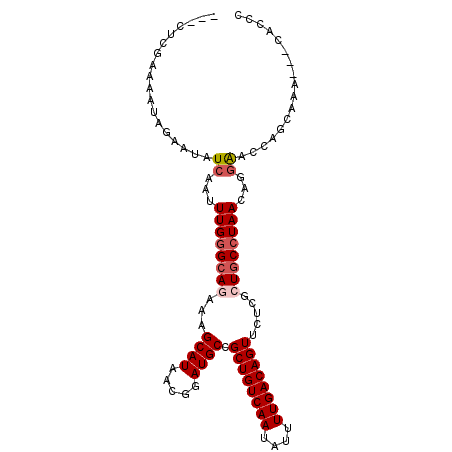
| Location | 19,136,744 – 19,136,844 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -20.44 |
| Energy contribution | -21.13 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19136744 100 - 20766785 ---CUCGAAAAUAGAAUAUCAAUUUGGGCAGAAAGCAUAACGGAUGCCGCUGUCAAUAUUUUGACAGUUCUCGCUGCCUAACAGGAACCAGCAAACAACACCC ---...............((...((((((((...((((.....)))).((((((((....)))))))).....))))))))...))................. ( -25.30) >DroSec_CAF1 20802 97 - 1 ---CUCGAAAAUAGAAUAUCAAUUUGGGCAGAAAGCAUAACGGAUGCCGCUGUCAAUAUUUUGACAGUUCUCGCUGCCUAACAGGAACCGGCAAG---CACCC ---...............((...((((((((...((((.....)))).((((((((....)))))))).....))))))))...))...((....---..)). ( -25.40) >DroEre_CAF1 25416 91 - 1 ---CUCCAAAAUAGAAUAUCAAUUUGGGCAGCAGGCAUAACGGAUGCCGCUGUCAAUAUUUUGACAGUUCUCGCUGCCUAACAGGAACCAGCAA--------- ---.(((......(.....)...(((((((((.(((((.....)))))((((((((....))))))))....)))))))))..)))........--------- ( -34.20) >DroWil_CAF1 41861 94 - 1 CCAGUUAAAAAUAGAAUAUCAAUUUGGGCAAAAAGCAUAACGGAUGCGGCUGUCAAUAUUUUGACAGUUCUCCCUACAUAAAGAGGAGCCCCAC--------- .........................((((.....((((.....))))(((((((((....))))))))).(((((......)).)))))))...--------- ( -24.20) >DroYak_CAF1 26714 100 - 1 ---CUCGAAAAUAGAAUAUCAAUUUGGGCAGAAAGCAUAACGGAUGCCGCUGUCAAUAUUUUGACAGUUCUCGCUGCCUAACAGGAACCAGCGAAACACACCC ---...............((...((((((((...((((.....)))).((((((((....)))))))).....))))))))...))................. ( -25.30) >DroAna_CAF1 29143 103 - 1 GCAUUCAAAAAUAGAAUAUCAAUUUGGGCAAAAAGCAUAACGGAUGCCGCUGUCAAUAUUUUGACAGUUCUCGCUGCCUAACGCAAACCACCGAACCACCGCC ((((((.......))))......(((((((....((((.....)))).((((((((....))))))))......))))))).))................... ( -24.10) >consensus ___CUCGAAAAUAGAAUAUCAAUUUGGGCAGAAAGCAUAACGGAUGCCGCUGUCAAUAUUUUGACAGUUCUCGCUGCCUAACAGGAACCAGCAAA___CACCC ..................((...((((((((...((((.....)))).((((((((....)))))))).....))))))))...))................. (-20.44 = -21.13 + 0.69)


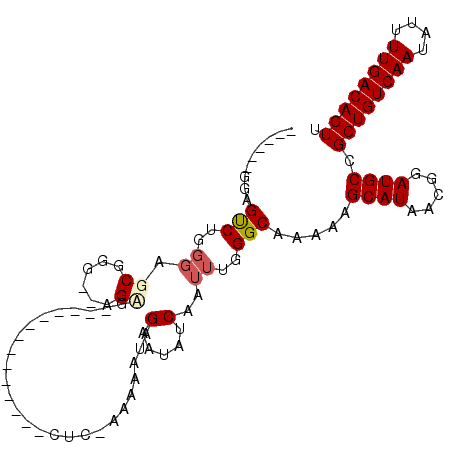
| Location | 19,136,778 – 19,136,869 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -24.29 |
| Consensus MFE | -13.81 |
| Energy contribution | -14.28 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19136778 91 - 20766785 -UCGGAGGAGCCUGGGAGCGGG--AGUG--------------CUC-GAAAAUAGAAUAUCAAUUUGGGCAGAAAGCAUAACGGAUGCCGCUGUCAAUAUUUUGACAGUU -.........((((....))))--..((--------------(((-(((.............))))))))....((((.....)))).((((((((....)))))))). ( -24.42) >DroPse_CAF1 29556 89 - 1 ------GGAGUCUGGGAGCGAGCGAGAA--------------CUCUUAAAAUAGAAUAUCAAUUUGGGCAAAAAGCAUAACGGAUGCCGCUGUCAAUAUUUUGACAGUU ------((.(((((..((((((......--------------)))........(..((......))..).....)).)..))))).))((((((((....)))))))). ( -21.60) >DroEre_CAF1 25441 86 - 1 ------GGAGCCUGGGAGCGGC--AGUG--------------CUC-CAAAAUAGAAUAUCAAUUUGGGCAGCAGGCAUAACGGAUGCCGCUGUCAAUAUUUUGACAGUU ------...((((.((((((..--..))--------------)))-).((((.(.....).))))))))....(((((.....)))))((((((((....)))))))). ( -30.80) >DroWil_CAF1 41886 78 - 1 ------GAAGUC-------G-----------------CCCCAGUU-AAAAAUAGAAUAUCAAUUUGGGCAAAAAGCAUAACGGAUGCGGCUGUCAAUAUUUUGACAGUU ------......-------(-----------------(((.((((-..............)))).)))).....((((.....))))(((((((((....))))))))) ( -20.84) >DroAna_CAF1 29177 106 - 1 GUCAGAAAAGUCUGGGAGCGGG--AGCGGGAGCAGCAACGCAUUC-AAAAAUAGAAUAUCAAUUUGGGCAAAAAGCAUAACGGAUGCCGCUGUCAAUAUUUUGACAGUU (((((((.(...(((.((((..--.((....))......((((((-..((((.(.....).))))..((.....)).....)))))))))).))).).))))))).... ( -26.50) >DroPer_CAF1 29643 89 - 1 ------GGAGUCUGGGAGCGAGCGAGAA--------------CUCUUAAAAUAGAAUAUCAAUUUGGGCAAAAAGCAUAACGGAUGCCGCUGUCAAUAUUUUGACAGUU ------((.(((((..((((((......--------------)))........(..((......))..).....)).)..))))).))((((((((....)))))))). ( -21.60) >consensus ______GGAGUCUGGGAGCGGG__AGAG______________CUC_AAAAAUAGAAUAUCAAUUUGGGCAAAAAGCAUAACGGAUGCCGCUGUCAAUAUUUUGACAGUU .........(((..((.((......))..........................(.....)..))..))).....((((.....)))).((((((((....)))))))). (-13.81 = -14.28 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:19 2006