
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,130,669 – 19,130,760 |
| Length | 91 |
| Max. P | 0.996336 |

| Location | 19,130,669 – 19,130,760 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -26.06 |
| Consensus MFE | -14.28 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
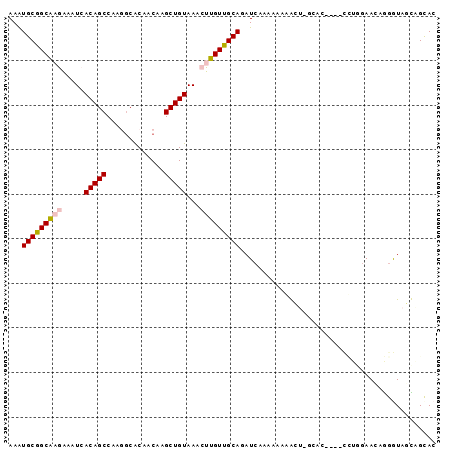
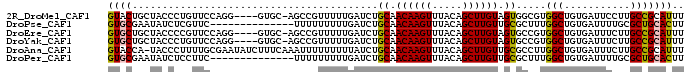
>2R_DroMel_CAF1 19130669 91 + 20766785 AAAUGCGGCAAGGAAUCACAGCCACGCCACUACAAGCUGUAAACUUGUUGCAGAUCAAAAACGGCU-GCAC----CCUGGAACAGGGUAGCAGUAC ...(((((((((.....(((((.............)))))...)))))))))...........(((-((((----((((...)))))).))))).. ( -33.02) >DroPse_CAF1 23394 82 + 1 AAGUGCAGCGCAAAAUCACAGCCAAAGCGCAACAAGCUGUAAACUUGUUGCAGAUCAAAAAAAAA--------------GAACGAGAUAUUCGCAC .......(((.((.(((...((....))((((((((.......))))))))..............--------------......))).))))).. ( -17.90) >DroEre_CAF1 19493 91 + 1 AAAUGCGGCAAGAAAUCACAGCCACGGCACUACAAGCUGUAAACUUGUUGCAGAUCAAAAACGGCU-GCAC----CCUGGAACGGGGUAGCAGCAC ...(((((((((.....(((((...(......)..)))))...)))))))))...........(((-((((----((((...)))))).))))).. ( -33.70) >DroYak_CAF1 20442 91 + 1 AAAUGCGGCAAGAAAUCACAGCCACGGCACUACAAGCUGUAAACUUGUUGCAGAUCAAAAACGGCU-GCAC----CCUGGAACAGGGUAGCAGCAC ...(((((((((.....(((((...(......)..)))))...)))))))))...........(((-((((----((((...)))))).))))).. ( -34.40) >DroAna_CAF1 22615 95 + 1 AAAUGCGGCAAGAAAUCACAGCCAAGGCGCAACAAGCUGUAAACUUGUUGCAGAUAAAAAAAAAUUUGAAAGAUAUUCGCAAAAGGGUA-UGGUAC ...(((((.......(((..((....))((((((((.......))))))))...............))).......)))))........-...... ( -19.34) >DroPer_CAF1 23412 82 + 1 AAGUGCAGCGCAAAAUCACAGCCAAAGCGCAACAAGCUGUAAACUUGUUGCAGAUCAAAAAAAAA--------------GAAGGAGAUAUUCGCAC ..((((........(((...((....))((((((((.......)))))))).)))..........--------------(((.......))))))) ( -18.00) >consensus AAAUGCGGCAAGAAAUCACAGCCAAGGCACAACAAGCUGUAAACUUGUUGCAGAUCAAAAAAAACU_GCAC____CCUGGAACAGGGUAGCAGCAC ...(((((((((.....(((((.............)))))...)))))))))............................................ (-14.28 = -14.50 + 0.22)


| Location | 19,130,669 – 19,130,760 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -13.19 |
| Energy contribution | -12.83 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
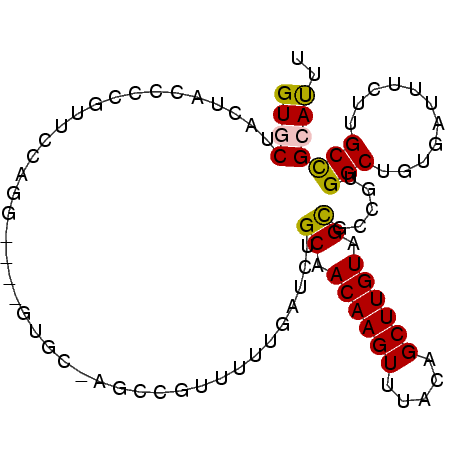
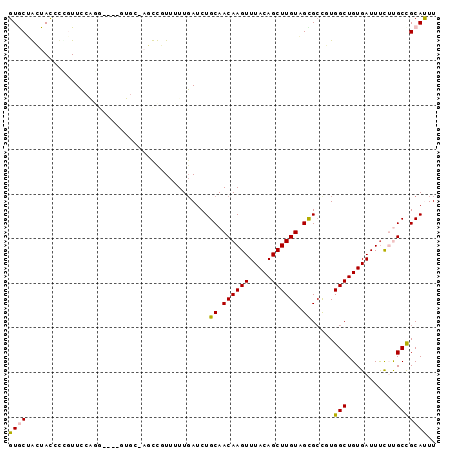
>2R_DroMel_CAF1 19130669 91 - 20766785 GUACUGCUACCCUGUUCCAGG----GUGC-AGCCGUUUUUGAUCUGCAACAAGUUUACAGCUUGUAGUGGCGUGGCUGUGAUUCCUUGCCGCAUUU ...((((.((((((...))))----))))-))...........(..(.((((((.....)))))).)..).(((((...........))))).... ( -30.70) >DroPse_CAF1 23394 82 - 1 GUGCGAAUAUCUCGUUC--------------UUUUUUUUUGAUCUGCAACAAGUUUACAGCUUGUUGCGCUUUGGCUGUGAUUUUGCGCUGCACUU (((((((.(((.(....--------------..............(((((((((.....)))))))))((....)).).))))))))))....... ( -24.10) >DroEre_CAF1 19493 91 - 1 GUGCUGCUACCCCGUUCCAGG----GUGC-AGCCGUUUUUGAUCUGCAACAAGUUUACAGCUUGUAGUGCCGUGGCUGUGAUUUCUUGCCGCAUUU (.(((((.((((.......))----))))-))))..........(((..((((.((((((((...........))))))))...))))..)))... ( -30.10) >DroYak_CAF1 20442 91 - 1 GUGCUGCUACCCUGUUCCAGG----GUGC-AGCCGUUUUUGAUCUGCAACAAGUUUACAGCUUGUAGUGCCGUGGCUGUGAUUUCUUGCCGCAUUU (.(((((.((((((...))))----))))-))))..........(((..((((.((((((((...........))))))))...))))..)))... ( -33.50) >DroAna_CAF1 22615 95 - 1 GUACCA-UACCCUUUUGCGAAUAUCUUUCAAAUUUUUUUUUAUCUGCAACAAGUUUACAGCUUGUUGCGCCUUGGCUGUGAUUUCUUGCCGCAUUU ......-........((((........(((...............(((((((((.....)))))))))((....))..)))........))))... ( -20.69) >DroPer_CAF1 23412 82 - 1 GUGCGAAUAUCUCCUUC--------------UUUUUUUUUGAUCUGCAACAAGUUUACAGCUUGUUGCGCUUUGGCUGUGAUUUUGCGCUGCACUU (((((((.(((......--------------..............(((((((((.....)))))))))((....))...))))))))))....... ( -23.30) >consensus GUGCUACUACCCCGUUCCAGG____GUGC_AGCCGUUUUUGAUCUGCAACAAGUUUACAGCUUGUAGCGCCGUGGCUGUGAUUUCUUGCCGCAUUU ((((.........................................((.((((((.....)))))).)).....(((...........))))))).. (-13.19 = -12.83 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:17 2006