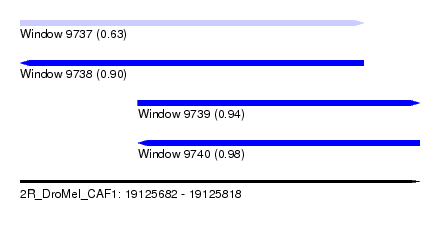
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,125,682 – 19,125,818 |
| Length | 136 |
| Max. P | 0.978013 |

| Location | 19,125,682 – 19,125,799 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -17.28 |
| Energy contribution | -17.08 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.633076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19125682 117 + 20766785 ACAAACGCAGCUCUUAAGAUGUAUUUAUGGCGCCACUGAAAGCUUUUCAAGCUUUCGCUUGCAUUUCCUAACGACCAGCUUUUUAUACCCUUAAAAGGGGGUAUUACGGAAUUAGUA ......(((((................((....))..(((((((.....))))))))).)))..((((.....(((..(((((((......))))))).))).....))))...... ( -25.90) >DroSec_CAF1 9925 116 + 1 ACAAACACAUCUCUUAAUAUGUAUUUAUGGCGCCACUGAAAGCUUUUCAAGCUUUCGCUUGCAUUUCCUAAGCACUAGCUUUUUAUACCCUUAA-AGGGGGUAUUACACAAUUAGUA .............(((((.((((.....(((......(((((((.....)))))))(((((.......)))))....)))....(((((((...-..)))))))))))..))))).. ( -24.40) >DroSim_CAF1 10040 116 + 1 ACAAACGCAGCUCUUAAUAUUUAUUUAUGGCGCCACUGAAAGCUUUUCAAGCUUUCGCUUGCAUUUCCUAAGCACUAGCUUUUUAUACCCUUAA-AGGGAGUAUUAUACAAUUAGUA ......((.(((..(((.......))).)))((...((((((((.....))))))))...)).........))(((((......(((((((...-.))).)))).......))))). ( -22.52) >DroEre_CAF1 14700 114 + 1 ACCAACGCAGCUCUAAAUAUGUACUUAUGGCGCCACCAAAAGCUUUUCAAGCUUUCGCUUGCAUUUU-UAACCUCUAGCUUUU-AUACCCUUAA-CUAGGGUAUUACACAAUAAGCG .....(((((((.((((.(((((.....((((......((((((.....))))))))))))))).))-))......))))...-(((((((...-..)))))))..........))) ( -23.70) >DroYak_CAF1 15352 109 + 1 ACCAACGCAGCUCUAAUUAUGUAUUUAUGGCGCCACCAAAAGCUUUUCAAGCUUUCGUAUGCAUUUU-------CCAGCUUUUGAUACCCUUAA-UGAGGGUAUUACACAUUUAGUA ......((((((......((((((...(((.....)))((((((.....))))))...))))))...-------..))))..((((((((((..-.))))))))))........)). ( -23.40) >consensus ACAAACGCAGCUCUUAAUAUGUAUUUAUGGCGCCACUGAAAGCUUUUCAAGCUUUCGCUUGCAUUUCCUAACCACUAGCUUUUUAUACCCUUAA_AGGGGGUAUUACACAAUUAGUA ......((.......................((...((((((((.....))))))))...))......................(((((((......)))))))..........)). (-17.28 = -17.08 + -0.20)

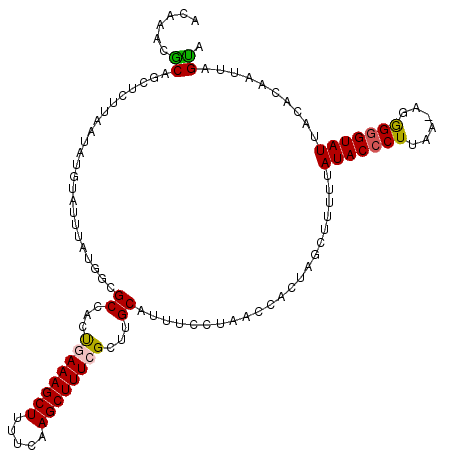
| Location | 19,125,682 – 19,125,799 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.50 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19125682 117 - 20766785 UACUAAUUCCGUAAUACCCCCUUUUAAGGGUAUAAAAAGCUGGUCGUUAGGAAAUGCAAGCGAAAGCUUGAAAAGCUUUCAGUGGCGCCAUAAAUACAUCUUAAGAGCUGCGUUUGU ..(((((.((((.((((((........)))))).....)).))..))))).(((((((.(((((((((.....))))))).(((....)))...............))))))))).. ( -30.10) >DroSec_CAF1 9925 116 - 1 UACUAAUUGUGUAAUACCCCCU-UUAAGGGUAUAAAAAGCUAGUGCUUAGGAAAUGCAAGCGAAAGCUUGAAAAGCUUUCAGUGGCGCCAUAAAUACAUAUUAAGAGAUGUGUUUGU .............((((((...-....)))))).....((((.((...(((.....(((((....))))).....))).)).))))...((((((((((........)))))))))) ( -32.30) >DroSim_CAF1 10040 116 - 1 UACUAAUUGUAUAAUACUCCCU-UUAAGGGUAUAAAAAGCUAGUGCUUAGGAAAUGCAAGCGAAAGCUUGAAAAGCUUUCAGUGGCGCCAUAAAUAAAUAUUAAGAGCUGCGUUUGU .............((((((...-....))))))...((((....))))...(((((((.(((((((((.....))))))).(((....)))...............))))))))).. ( -26.60) >DroEre_CAF1 14700 114 - 1 CGCUUAUUGUGUAAUACCCUAG-UUAAGGGUAU-AAAAGCUAGAGGUUA-AAAAUGCAAGCGAAAGCUUGAAAAGCUUUUGGUGGCGCCAUAAGUACAUAUUUAGAGCUGCGUUGGU .((((((.((((.(((((((..-...)))))))-....((((((((((.-......(((((....)))))...)))))))))).)))).))))))...................... ( -36.20) >DroYak_CAF1 15352 109 - 1 UACUAAAUGUGUAAUACCCUCA-UUAAGGGUAUCAAAAGCUGG-------AAAAUGCAUACGAAAGCUUGAAAAGCUUUUGGUGGCGCCAUAAAUACAUAAUUAGAGCUGCGUUGGU ..((((.(((((((((((((..-...))))))).......(((-------....(((...((((((((.....))))))))...))))))....)))))).))))............ ( -29.30) >consensus UACUAAUUGUGUAAUACCCCCU_UUAAGGGUAUAAAAAGCUAGUGCUUAGGAAAUGCAAGCGAAAGCUUGAAAAGCUUUCAGUGGCGCCAUAAAUACAUAUUAAGAGCUGCGUUUGU .............((((((........))))))..................(((((((.(((((((((.....))))))).(((....)))...............))))))))).. (-21.26 = -21.50 + 0.24)

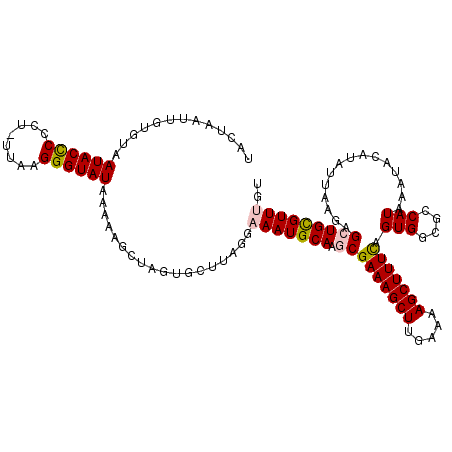
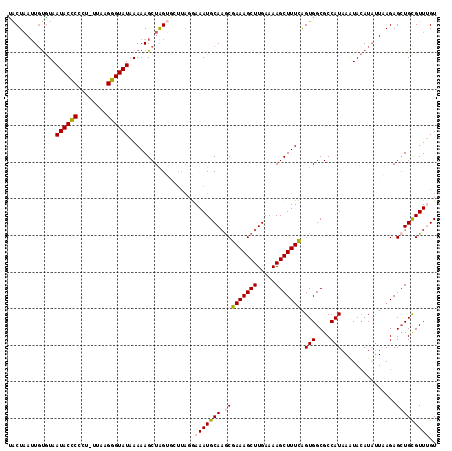
| Location | 19,125,722 – 19,125,818 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -13.42 |
| Energy contribution | -14.10 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19125722 96 + 20766785 AGCUUUUCAAGCUUUCGCUUGCAUUUCCUAACGACCAGCUUUUUAUACCCUUAAAAGGGGGUAUUACGGAAUUAGUAAGAGCUUUACAAAGCAGUG .(((((..(((((((..((.....((((.....(((..(((((((......))))))).))).....))))..))..)))))))...))))).... ( -27.40) >DroSec_CAF1 9965 95 + 1 AGCUUUUCAAGCUUUCGCUUGCAUUUCCUAAGCACUAGCUUUUUAUACCCUUAA-AGGGGGUAUUACACAAUUAGUAAGAGCUUUGCAACGCAGUG .((....(((((....)))))..........(((..(((((((.(((((((...-..)))))))............))))))).)))...)).... ( -24.52) >DroSim_CAF1 10080 95 + 1 AGCUUUUCAAGCUUUCGCUUGCAUUUCCUAAGCACUAGCUUUUUAUACCCUUAA-AGGGAGUAUUAUACAAUUAGUAAUAGCUUUGCAACGUGGUG ((((.....)))).(((((((((....(((...(((((......(((((((...-.))).)))).......)))))..)))...))))).)))).. ( -18.12) >DroEre_CAF1 14740 83 + 1 AGCUUUUCAAGCUUUCGCUUGCAUUUU-UAACCUCUAGCUUUU-AUACCCUUAA-CUAGGGUAUUACACAAUAAGCGAGAA--------AGCAG-- .(((((((((((....)))))......-.........((((..-(((((((...-..)))))))........))))..)))--------)))..-- ( -21.90) >DroYak_CAF1 15392 88 + 1 AGCUUUUCAAGCUUUCGUAUGCAUUUU-------CCAGCUUUUGAUACCCUUAA-UGAGGGUAUUACACAUUUAGUAAGAACUUUGUAAAGCAAUA .(((((.((((..(((.(((((.....-------...))...((((((((((..-.))))))))))........))).))).)))).))))).... ( -20.20) >consensus AGCUUUUCAAGCUUUCGCUUGCAUUUCCUAACCACUAGCUUUUUAUACCCUUAA_AGGGGGUAUUACACAAUUAGUAAGAGCUUUGCAAAGCAGUG .((....(((((....)))))...............(((((((.(((((((......)))))))............))))))).......)).... (-13.42 = -14.10 + 0.68)

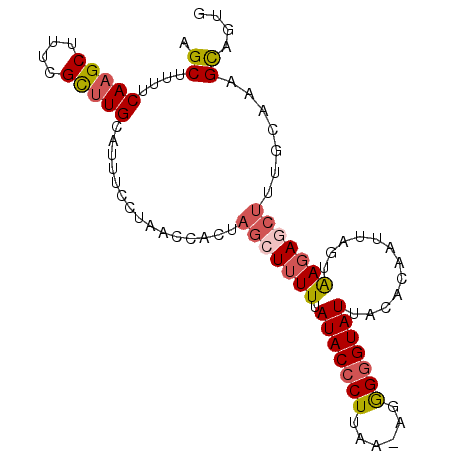
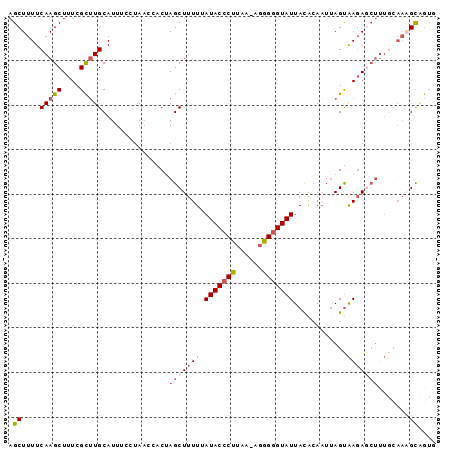
| Location | 19,125,722 – 19,125,818 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -15.48 |
| Energy contribution | -16.84 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.978013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19125722 96 - 20766785 CACUGCUUUGUAAAGCUCUUACUAAUUCCGUAAUACCCCCUUUUAAGGGUAUAAAAAGCUGGUCGUUAGGAAAUGCAAGCGAAAGCUUGAAAAGCU ....(((((((((.....))))...((((...((((((........))))))....(((.....))).))))...(((((....))))).))))). ( -24.90) >DroSec_CAF1 9965 95 - 1 CACUGCGUUGCAAAGCUCUUACUAAUUGUGUAAUACCCCCU-UUAAGGGUAUAAAAAGCUAGUGCUUAGGAAAUGCAAGCGAAAGCUUGAAAAGCU ....((...(((.((((..(((.......)))((((((...-....))))))....))))..)))..........(((((....)))))....)). ( -24.70) >DroSim_CAF1 10080 95 - 1 CACCACGUUGCAAAGCUAUUACUAAUUGUAUAAUACUCCCU-UUAAGGGUAUAAAAAGCUAGUGCUUAGGAAAUGCAAGCGAAAGCUUGAAAAGCU ..((.....(((.((((..(((.....)))..((((((...-....))))))....))))..)))...)).....(((((....)))))....... ( -21.60) >DroEre_CAF1 14740 83 - 1 --CUGCU--------UUCUCGCUUAUUGUGUAAUACCCUAG-UUAAGGGUAU-AAAAGCUAGAGGUUA-AAAAUGCAAGCGAAAGCUUGAAAAGCU --..(((--------((...((((...((...(((((((..-...)))))))-....))...))))..-......(((((....))))).))))). ( -25.10) >DroYak_CAF1 15392 88 - 1 UAUUGCUUUACAAAGUUCUUACUAAAUGUGUAAUACCCUCA-UUAAGGGUAUCAAAAGCUGG-------AAAAUGCAUACGAAAGCUUGAAAAGCU ....(((((.((((((....)))...(((((.(((((((..-...))))))).....((...-------.....))))))).....))).))))). ( -18.20) >consensus CACUGCUUUGCAAAGCUCUUACUAAUUGUGUAAUACCCCCU_UUAAGGGUAUAAAAAGCUAGUGCUUAGGAAAUGCAAGCGAAAGCUUGAAAAGCU ....(((((....((((..(((.....)))..((((((........))))))....))))...............(((((....))))).))))). (-15.48 = -16.84 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:15 2006