
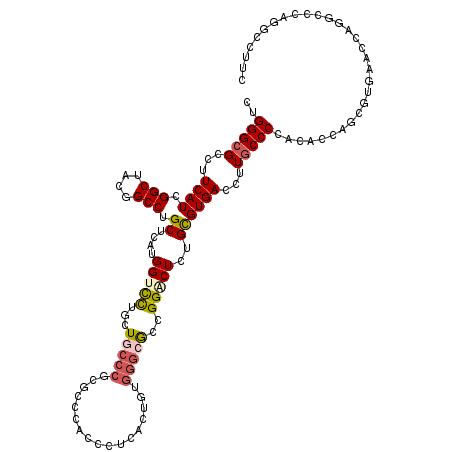
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,115,863 – 19,116,063 |
| Length | 200 |
| Max. P | 0.940190 |

| Location | 19,115,863 – 19,115,983 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -51.42 |
| Consensus MFE | -23.41 |
| Energy contribution | -23.91 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
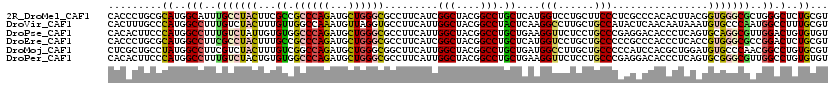
>2R_DroMel_CAF1 19115863 120 - 20766785 CUGGGCGCCUUCAUCGGCUACGGCCUGCUCAUGGUCCUGCUUCCCUCGCCCACACUUACGGUGGGCGCUGGGCUCUGCGUGACCUUGCCCCACACCAGCGUAACCACGGGCCAGGCCCUC ..(((((((......)))...((((((.....((((.(((..(((.(((((((.......)))))))..)))....))).))))..((.........)).......))))))..)))).. ( -51.80) >DroPse_CAF1 7822 120 - 1 CUGGGCGCCUUCAUUGGCUACGGCCUGCUGAAGGUUCUCCUGCCCGAGGACACCCUCAGUGCAGGCGUUGGACUGUGUGUGACCUUACCCCACAACAGCAUUAGCCAUGCUCAGGCCUUU (((((((((((((..(((....)))...)))))))...((((((.((((....)))).).))))).(((((.((((.((((.........))))))))..)))))...))))))...... ( -47.50) >DroEre_CAF1 4974 120 - 1 CUGGGCGCCUUCAUCGGCUACGGCCUGCUCAUGGUCCUGCUGCCCCCGCCCACCCUCACCGUGGGCGCCGGACUCUGCGUGACCUUGCCCCAUGCCACCGUGAACGUGGGCCAGGCCUUC ..(((((((......)))...((((..((((((((.......((..(((((((.......)))))))..)).....(((((.........))))).)))))))..)..))))..)))).. ( -50.30) >DroYak_CAF1 5341 120 - 1 CUGGGCGCCUUCAUCGGCUACGGCCUGCUCAUGGUCCUGCUCCCCGGGCCCACACUCACCGUGGGCGCCGGGCUCUGCGUGACCUUGCCCCACGCCACCGUGACCCCGGCCCAGGCCUUC (((((((((......)))..(((...((....((((.(((..(((((((((((.......)))))).)))))....))).))))..))..((((....))))...)))))))))...... ( -57.20) >DroMoj_CAF1 6744 120 - 1 CUGGGCGGCUUCAUUGGCUACGGCCUGCUGAUGGCCUUGCUGCCCCCAUCCACGCUGGAUGUGCCCAACGGCCUGUGCGUGACCCUGCCCCACUCCGAGGUCAACAAUCUGCAGGCCUUC .((((((((..((..((((((((....))).))))).))..)))..(((((.....))))).)))))..(((((((...(((((.((........)).))))).......)))))))... ( -50.50) >DroPer_CAF1 7834 120 - 1 CUGGGCGCCUUCAUUGGCUACGGCCUGCUGAAGGUUCUCCUGCCCGAGGACACCCUCAGUGCGGGCGUUGGCCUGUGUGUGACCUUGCCCCACAGCAGCAUUAGCCAUGCUCAGGCCUUU ..(((.(((((((..(((....)))...))))))).)))..((((((((....)))).....))))...((((((.(((((....(((......)))((....))))))).))))))... ( -51.20) >consensus CUGGGCGCCUUCAUCGGCUACGGCCUGCUCAUGGUCCUGCUGCCCGCGCCCACCCUCACUGUGGGCGCCGGACUCUGCGUGACCUUGCCCCACACCAGCGUGAACCAGGCCCAGGCCUUC ..(((((...((((.(((....))).((....(((((...(((((.................)))))..)))))..))))))...))))).............................. (-23.41 = -23.91 + 0.50)

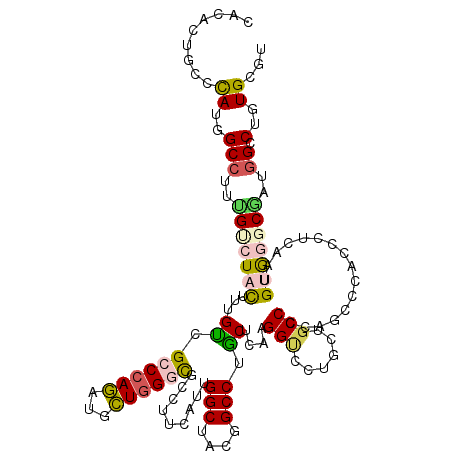
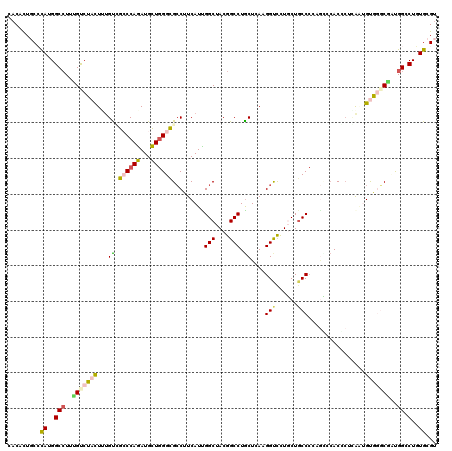
| Location | 19,115,903 – 19,116,023 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -51.35 |
| Consensus MFE | -22.42 |
| Energy contribution | -23.53 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
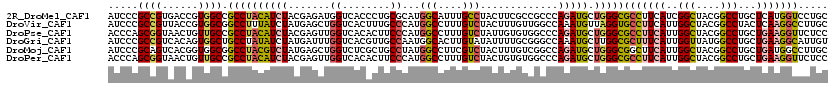
>2R_DroMel_CAF1 19115903 120 + 20766785 ACGCAGAGCCCAGCGCCCACCGUAAGUGUGGGCGAGGGAAGCAGGACCAUGAGCAGGCCGUAGCCGAUGAAGGCGCCCAGCAUCUGGGCGGCGAAGUAGGCAAAUGCCAUGCGCAGGGUG ..((....(((..((((((((....).))))))).)))..))...(((.((.(((.(((...(((......)))((((((...)))))))))......(((....))).))).)).))). ( -58.00) >DroVir_CAF1 176 120 + 1 ACGCAAAGGCCAUUGGGCACAUUUAUUGUUGAGUAUGGCAGCAAGGCCUUGAGUAGGCCGUAGCCAAUGAAGGCACCUAACAUUUGGCCAACAAAGUAGACAAAGGCCAUGGGCAAAGUG ..((...((((.((((((((.....((((((.......))))))(((((.....))))))).)))..((..(((............)))..)).......))).))))....))...... ( -36.20) >DroPse_CAF1 7862 120 + 1 ACACACAGUCCAACGCCUGCACUGAGGGUGUCCUCGGGCAGGAGAACCUUCAGCAGGCCGUAGCCAAUGAAGGCGCCCAGCAUCUGGGCCACACAAUAGACAAAGGCCAUGGGAAGUGUG .(((((..((((.(.(((((.((((((....))))))))))).)...........((((...(((......)))((((((...))))))...............)))).))))..))))) ( -53.70) >DroEre_CAF1 5014 120 + 1 ACGCAGAGUCCGGCGCCCACGGUGAGGGUGGGCGGGGGCAGCAGGACCAUGAGCAGGCCGUAGCCGAUGAAGGCGCCCAGCAUCUGGGCGGCAAAGUAGGCGAAGGCCAUGCGCAGGGUG ..((...((((..(((((((.......))))))).)))).))...(((.((.(((((((...(((.((...(.(((((((...))))))).)...)).)))...)))).))).)).))). ( -59.80) >DroMoj_CAF1 6784 120 + 1 ACGCACAGGCCGUUGGGCACAUCCAGCGUGGAUGGGGGCAGCAAGGCCAUCAGCAGGCCGUAGCCAAUGAAGCCGCCCAGCAUCUGGCCGACAAAGUAGACGAAGGCCAUAGGCAGCGAG .(((....((((((((((.(((((.....)))))..(((.((..((((.......)))))).))).........)))))))...(((((..(.........)..)))))..))).))).. ( -50.90) >DroPer_CAF1 7874 120 + 1 ACACACAGGCCAACGCCCGCACUGAGGGUGUCCUCGGGCAGGAGAACCUUCAGCAGGCCGUAGCCAAUGAAGGCGCCCAGCAUCUGGGCCACACAGUAGACAAAGGCCAUGGGAAGUGUG .(((((.(((....)))(.((((((((((.((((.....))))..))))))))..((((...(((......)))((((((...))))))...............)))).)).)..))))) ( -49.50) >consensus ACGCACAGGCCAACGCCCACACUGAGGGUGGACGAGGGCAGCAGGACCAUCAGCAGGCCGUAGCCAAUGAAGGCGCCCAGCAUCUGGGCGACAAAGUAGACAAAGGCCAUGGGCAGGGUG .(((...(.((.((((((.......))))))....)).)................((((...(((......)))((((((...))))))...............)))).........))) (-22.42 = -23.53 + 1.12)


| Location | 19,115,903 – 19,116,023 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -48.75 |
| Consensus MFE | -24.42 |
| Energy contribution | -24.73 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19115903 120 - 20766785 CACCCUGCGCAUGGCAUUUGCCUACUUCGCCGCCCAGAUGCUGGGCGCCUUCAUCGGCUACGGCCUGCUCAUGGUCCUGCUUCCCUCGCCCACACUUACGGUGGGCGCUGGGCUCUGCGU .(((.((.(((.(((....)))......(((((((((...))))))(((......)))...))).))).)).)))...((..(((.(((((((.......)))))))..)))....)).. ( -51.60) >DroVir_CAF1 176 120 - 1 CACUUUGCCCAUGGCCUUUGUCUACUUUGUUGGCCAAAUGUUAGGUGCCUUCAUUGGCUACGGCCUACUCAAGGCCUUGCUGCCAUACUCAACAAUAAAUGUGCCCAAUGGCCUUUGCGU ......((....((((.(((..((((((((((.....(((.((((((((......))).))(((((.....)))))...))).)))......))))))).)))..))).))))...)).. ( -36.50) >DroPse_CAF1 7862 120 - 1 CACACUUCCCAUGGCCUUUGUCUAUUGUGUGGCCCAGAUGCUGGGCGCCUUCAUUGGCUACGGCCUGCUGAAGGUUCUCCUGCCCGAGGACACCCUCAGUGCAGGCGUUGGACUGUGUGU (((((.(((.(((..................((((((...))))))(((((((..(((....)))...)))))))...((((((.((((....)))).).)))))))).)))..))))). ( -51.70) >DroEre_CAF1 5014 120 - 1 CACCCUGCGCAUGGCCUUCGCCUACUUUGCCGCCCAGAUGCUGGGCGCCUUCAUCGGCUACGGCCUGCUCAUGGUCCUGCUGCCCCCGCCCACCCUCACCGUGGGCGCCGGACUCUGCGU .(((.((.(((.((((...(((......(.(((((((...))))))).)......)))...))))))).)).)))...((.(((..(((((((.......)))))))..)).)...)).. ( -50.50) >DroMoj_CAF1 6784 120 - 1 CUCGCUGCCUAUGGCCUUCGUCUACUUUGUCGGCCAGAUGCUGGGCGGCUUCAUUGGCUACGGCCUGCUGAUGGCCUUGCUGCCCCCAUCCACGCUGGAUGUGCCCAACGGCCUGUGCGU ..(((.(((..(((((..((.......))..)))))...(.((((((((..((..((((((((....))).))))).))..)))..(((((.....))))).))))).))))....))). ( -48.30) >DroPer_CAF1 7874 120 - 1 CACACUUCCCAUGGCCUUUGUCUACUGUGUGGCCCAGAUGCUGGGCGCCUUCAUUGGCUACGGCCUGCUGAAGGUUCUCCUGCCCGAGGACACCCUCAGUGCGGGCGUUGGCCUGUGUGU (((((.......((((..(((((((((.(((((((((...))))))(((((((..(((....)))...)))))))..((((.....)))))))...))))..)))))..)))).))))). ( -53.90) >consensus CACACUGCCCAUGGCCUUUGUCUACUUUGUCGCCCAGAUGCUGGGCGCCUUCAUUGGCUACGGCCUGCUCAAGGUCCUGCUGCCCCAGCCCACCCUCAAUGUGGGCGAUGGCCUGUGCGU .........((..(((..(((((((...((.((((((...)))))).........(((....))).))....(((......)))................)))))))..)).)..))... (-24.42 = -24.73 + 0.31)
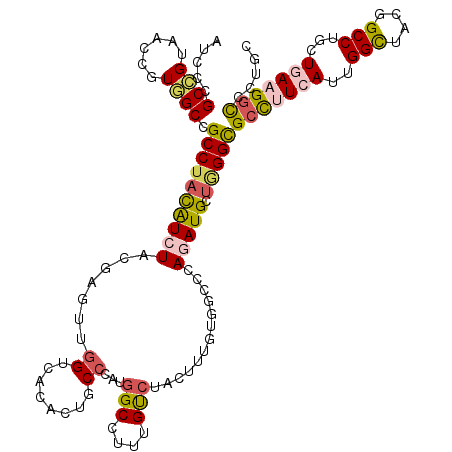
| Location | 19,115,943 – 19,116,063 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -43.06 |
| Consensus MFE | -25.71 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19115943 120 - 20766785 AUCCCGCCGUGACCGUGGCCGCCUACAUCUACGAGAUGGUCACCCUGCGCAUGGCAUUUGCCUACUUCGCCGCCCAGAUGCUGGGCGCCUUCAUCGGCUACGGCCUGCUCAUGGUCCUGC .....((((((((((((((((....((((.....))))........(((...(((....))).....)))(((((((...))))))).......))))))))).....)))))))..... ( -46.10) >DroVir_CAF1 216 120 - 1 AUCCCGCCGUUACCGUGGCGGCCUUUAUCUAUGAGCUGGUCACUUUGCCCAUGGCCUUUGUCUACUUUGUUGGCCAAAUGUUAGGUGCCUUCAUUGGCUACGGCCUACUCAAGGCCUUGC .....((((......))))((((((.....(((((..((.(((((.((...(((((...............)))))...)).)))))))))))).(((....))).....)))))).... ( -39.36) >DroPse_CAF1 7902 120 - 1 ACCCAGCGGUAACUGUUGCCGCCUACAUCUACGAGUUGGUCACACUUCCCAUGGCCUUUGUCUAUUGUGUGGCCCAGAUGCUGGGCGCCUUCAUUGGCUACGGCCUGCUGAAGGUUCUCC .(((((((....(((..(((((..(((...(((((..(((((.........))))))))))....)))))))).))).))))))).(((((((..(((....)))...)))))))..... ( -45.40) >DroGri_CAF1 7211 120 - 1 AUCCCGCCGUCACAGUGGCUGCCUAUAUCUAUGAUUUGGUCACGUUGCCAAUGGCACUUGUAUAUUUUGCGGGCCAAAUGCUUGGCGCUUUCAUUGGUUAUGGCCUGCUGAAGGCAUUGU .....(((....(((((((.(((...(((...)))..)))..(((.((((((((.....(((.....)))(.(((((....))))).)..)))))))).)))))).))))..)))..... ( -36.70) >DroMoj_CAF1 6824 120 - 1 AUCCCGCAGUCACGGUGGCGGCCUACGUCUAUGAGCUGGUCUCGCUGCCUAUGGCCUUCGUCUACUUUGUCGGCCAGAUGCUGGGCGGCUUCAUUGGCUACGGCCUGCUGAUGGCCUUGC ..((((((.((..((..((((((..(........)..)))..)))..))..(((((..((.......))..)))))))))).))).(((((((..(((....)))...))).)))).... ( -45.60) >DroPer_CAF1 7914 120 - 1 ACCCAGCGGUAACUGUUGCCGCCUACAUCUACGAGUUGGUCACACUUCCCAUGGCCUUUGUCUACUGUGUGGCCCAGAUGCUGGGCGCCUUCAUUGGCUACGGCCUGCUGAAGGUUCUCC .((((((((((.....)))).................((((((((.......(((....)))....)))))))).....)))))).(((((((..(((....)))...)))))))..... ( -45.20) >consensus AUCCCGCCGUAACCGUGGCCGCCUACAUCUACGAGUUGGUCACACUGCCCAUGGCCUUUGUCUACUUUGUGGCCCAGAUGCUGGGCGCCUUCAUUGGCUACGGCCUGCUGAAGGCCCUGC .....((((......)))).((((((((((.......((........))...(((....))).............))))).)))))(((((((..(((....)))...)))))))..... (-25.71 = -26.10 + 0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:09 2006