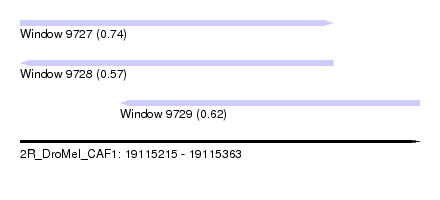
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,115,215 – 19,115,363 |
| Length | 148 |
| Max. P | 0.739264 |

| Location | 19,115,215 – 19,115,331 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.99 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -21.94 |
| Energy contribution | -22.73 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.739264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19115215 116 + 20766785 UUCUUUAUUUCUGAAGGCAAUUAGUGUUUUCACCAAGACGUUUAUCGUUUCAAGGGCUUAUGUAAGUGUGUGCACUUGGUUCGCCUUUUGAAACGUUCGAGCCACUUUGUGAAGAC .((((((....))))))........((((((((.(((..((((..((((((((((((..((.((((((....))))))))..))).)))))))))...))))..))).)))))))) ( -34.40) >DroEre_CAF1 4364 106 + 1 UUCUUUAUUACGGAAGGCAAUUAGUGUUUUCACCAAGUUGAUUAUCGUUCC-AAGGCUU----GAGU--GCACACUUGAU-CGCU-UUUGGAACGUUCGAG-CACUUUGUGAAGAC ..........(....).........((((((((.((((.......((((((-((((((.----.(((--....)))..).-.)).-)))))))))......-.)))).)))))))) ( -30.34) >DroYak_CAF1 4688 109 + 1 UUCUUUAUUACUGAAGGCAAUUGGUGUGUUCACCAAGUCGUUUAUGGUUUCAAAGGCUU----GAGUGUGUACACUUGAU-CGCC-AUCGAAAGGUUCGAG-CACUUUGUGAAGAC .(((((((....((((((....((((((..(((((((((...............)))))----).)))..))))))...(-((..-(((....))).))))-).))))))))))). ( -31.16) >consensus UUCUUUAUUACUGAAGGCAAUUAGUGUUUUCACCAAGUCGUUUAUCGUUUCAAAGGCUU____GAGUGUGUACACUUGAU_CGCC_UUUGAAACGUUCGAG_CACUUUGUGAAGAC .(((((......)))))........((((((((.((((.......((((((...(((......(((((....))))).....)))....))))))........)))).)))))))) (-21.94 = -22.73 + 0.78)

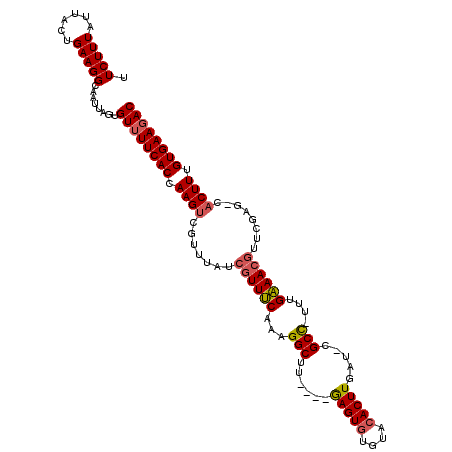
| Location | 19,115,215 – 19,115,331 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.99 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -17.63 |
| Energy contribution | -19.08 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19115215 116 - 20766785 GUCUUCACAAAGUGGCUCGAACGUUUCAAAAGGCGAACCAAGUGCACACACUUACAUAAGCCCUUGAAACGAUAAACGUCUUGGUGAAAACACUAAUUGCCUUCAGAAAUAAAGAA .((((........(((.....((((((((..(((.....(((((....)))))......))).)))))))).........((((((....))))))..)))..........)))). ( -26.17) >DroEre_CAF1 4364 106 - 1 GUCUUCACAAAGUG-CUCGAACGUUCCAAA-AGCG-AUCAAGUGUGC--ACUC----AAGCCUU-GGAACGAUAAUCAACUUGGUGAAAACACUAAUUGCCUUCCGUAAUAAAGAA ((.(((((.((((.-......((((((((.-.((.-....(((....--))).----..)).))-)))))).......)))).))))).))....(((((.....)))))...... ( -24.24) >DroYak_CAF1 4688 109 - 1 GUCUUCACAAAGUG-CUCGAACCUUUCGAU-GGCG-AUCAAGUGUACACACUC----AAGCCUUUGAAACCAUAAACGACUUGGUGAACACACCAAUUGCCUUCAGUAAUAAAGAA .((((.......((-((.(((..((((((.-(((.-....((((....)))).----..))).)))))).......(((.((((((....)))))))))..)))))))...)))). ( -23.20) >consensus GUCUUCACAAAGUG_CUCGAACGUUUCAAA_GGCG_AUCAAGUGUACACACUC____AAGCCUUUGAAACGAUAAACGACUUGGUGAAAACACUAAUUGCCUUCAGUAAUAAAGAA ((.(((((.((((........(((((((...(((......((((....)))).......)))..))))))).......)))).))))).))......................... (-17.63 = -19.08 + 1.45)

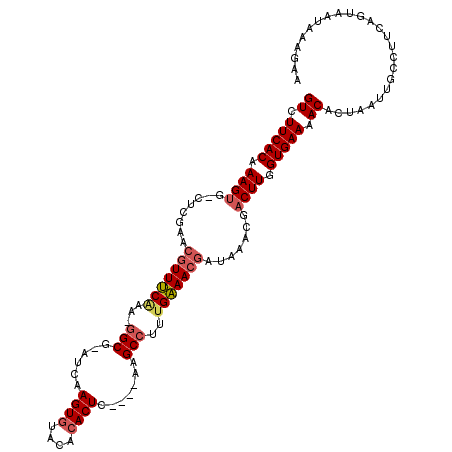
| Location | 19,115,252 – 19,115,363 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.44 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -20.37 |
| Energy contribution | -21.49 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19115252 111 - 20766785 UAAUGCAUUCGUUGUACGCUAAUGUUAGCAGUGUCUUCACAAAGUGGCUCGAACGUUUCAAAAGGCGAACCAAGUGCACACACUUACAUAAGCCCUUGAAACGAUAAACGU .......((((...(((((((....)))).(((....)))...)))...))))((((((((..(((.....(((((....)))))......))).))))))))........ ( -27.20) >DroEre_CAF1 4401 101 - 1 UAAUGCAUUCGUUGUACGCUAAUCUUAGCAGUGUCUUCACAAAGUG-CUCGAACGUUCCAAA-AGCG-AUCAAGUGUGC--ACUC----AAGCCUU-GGAACGAUAAUCAA .......((((..((((((((....)))).(((....)))...)))-).))))((((((((.-.((.-....(((....--))).----..)).))-))))))........ ( -26.50) >DroYak_CAF1 4725 104 - 1 UAAUGCAUUCGUUGUACGCUACUGUUAGCAGUGUCUUCACAAAGUG-CUCGAACCUUUCGAU-GGCG-AUCAAGUGUACACACUC----AAGCCUUUGAAACCAUAAACGA .......((((..((((((((....)))).(((....)))...)))-).))))..((((((.-(((.-....((((....)))).----..))).)))))).......... ( -25.70) >consensus UAAUGCAUUCGUUGUACGCUAAUGUUAGCAGUGUCUUCACAAAGUG_CUCGAACGUUUCAAA_GGCG_AUCAAGUGUACACACUC____AAGCCUUUGAAACGAUAAACGA .......((((...(((((((....)))).(((....)))...)))...))))(((((((...(((......((((....)))).......)))..)))))))........ (-20.37 = -21.49 + 1.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:05 2006