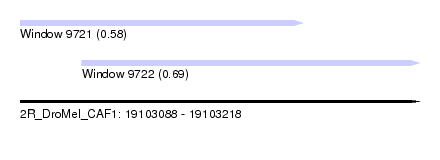
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,103,088 – 19,103,218 |
| Length | 130 |
| Max. P | 0.686469 |

| Location | 19,103,088 – 19,103,180 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 87.94 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -16.71 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19103088 92 + 20766785 GUACGCGAUAAAUGCGAAUGUACAUAUAUACGU---AUGUAUCUAGAUACGUAGUUCGAGUGUACGAGGUACACAUACGAGUGCCGAGUAUAAGU ((((((((((.(((((.((((....)))).)))---)).))))......((.....)).))))))..(((((........))))).......... ( -24.20) >DroSec_CAF1 45595 94 + 1 GUACGCGAUAAAUGCGAAUGUACAUAUCUAUAUGCGACGUAUCUAGAUACGUAGUUCGAGUGUACGAGGUACACAUACGAGUGC-GAGUAUAAGU ((((.((.......(((((.(((.((((((.((((...)))).)))))).)))))))).((((((...))))))...)).))))-.......... ( -26.10) >DroSim_CAF1 46461 94 + 1 GUACGCGAUAAAUGCGAAUGUACAUAUCUACAUGCGACGUAUCUAGAUACGUAGUUCGAGUGUACGAGGUACACAUACGAGUGC-GAGUAUAAGU ((((.((.......(((((.(((.((((((.((((...)))).)))))).)))))))).((((((...))))))...)).))))-.......... ( -26.10) >DroEre_CAF1 42116 85 + 1 GUACGCGAUAAAUGCGAAUGUACAUACAUGUCU---ACGUAUCUAGAUACGUAGUUCGAGUGUACGAGGUACACAUACGAGUG-------UAUGU ...((((.....)))).....((((((((..((---((((((....)))))))).(((.((((((...))))))...))))))-------))))) ( -26.80) >DroYak_CAF1 59553 85 + 1 GUACGCGAUAAAUGCGAAUGUACAUACACGUCU---AUGUAUCUAGAUACGUAGUUCGAGUGUACGAGGUACACAUACGAGUU-------UGAGU (((((((.....))))..(((((.(((((..((---((((((....)))))))).....)))))....)))))..))).....-------..... ( -21.40) >consensus GUACGCGAUAAAUGCGAAUGUACAUAUAUAUAU___ACGUAUCUAGAUACGUAGUUCGAGUGUACGAGGUACACAUACGAGUGC_GAGUAUAAGU ((((.((.......(((((.................((((((....)))))).))))).((((((...))))))...)).))))........... (-16.71 = -17.07 + 0.36)


| Location | 19,103,108 – 19,103,218 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -23.48 |
| Consensus MFE | -13.62 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19103108 110 + 20766785 UACAUAUAUACGU---AUGUAUCUAGAUACGUAGUUCGAGUGUACGAGGUACACAUACGAGUGCCGAGUAUA-AGUAUAAAUUGUAAAUCAGAUACGGCAUAAAUAUUGAACAC ........(((((---((.(....).)))))))((((((((((((...))))))......((((((.((...-..(((.....)))......)).)))))).....)))))).. ( -27.40) >DroSec_CAF1 45615 112 + 1 UACAUAUCUAUAUGCGACGUAUCUAGAUACGUAGUUCGAGUGUACGAGGUACACAUACGAGUGC-GAGUAUA-AGUAUAAAUUGUAAAUCAGAUACGGCAUAAAUAUUGAAAAC ..........(((((..(((((((.(((.((((.((((.((((((...))))))...)))))))-)..((((-(.......))))).)))))))))))))))............ ( -27.30) >DroSim_CAF1 46481 112 + 1 UACAUAUCUACAUGCGACGUAUCUAGAUACGUAGUUCGAGUGUACGAGGUACACAUACGAGUGC-GAGUAUA-AGUAUAAAUUGUAAAUCAGAUACGGCAUAAAUAUUGAACAC (((.((((((.((((...)))).)))))).)))((((((((((((...))))))......((((-..((((.-..(((.....)))......)))).)))).....)))))).. ( -26.10) >DroEre_CAF1 42136 103 + 1 UACAUACAUGUCU---ACGUAUCUAGAUACGUAGUUCGAGUGUACGAGGUACACAUACGAGUG-------UA-UGUAUAAAUUGUAAAUCAGAUACGGCAUAAAUAUUGAACAC (((((((((..((---((((((....)))))))).(((.((((((...))))))...))))))-------))-))))....(((((.......)))))................ ( -25.60) >DroYak_CAF1 59573 103 + 1 UACAUACACGUCU---AUGUAUCUAGAUACGUAGUUCGAGUGUACGAGGUACACAUACGAGUU-------UG-AGUAUAAAUUGUAAAUCAGAUACGGCAUAAAUAUUGAACAC ............(---((((((....)))))))((((((((((((...))))))...((.(((-------((-(.(((.....)))..)))))).)).........)))))).. ( -22.30) >DroAna_CAF1 41702 87 + 1 UAAAUACAUAUA-----UUUAUCUUUAUG-------UGAGUG--------CCACAUACAUGCA-------UACUAUAUAAAUUGUAAAUCAGAUACGGUAUAAAUAUUGAACAC ......((.(((-----(((((((.(((.-------((((((--------(.........)))-------)..((((.....))))..))).))).)).))))))))))..... ( -12.20) >consensus UACAUACAUAUAU___ACGUAUCUAGAUACGUAGUUCGAGUGUACGAGGUACACAUACGAGUG_______UA_AGUAUAAAUUGUAAAUCAGAUACGGCAUAAAUAUUGAACAC .................(((((((.(((...........((((((...)))))).(((((.....................))))).))))))))))................. (-13.62 = -13.98 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:58 2006