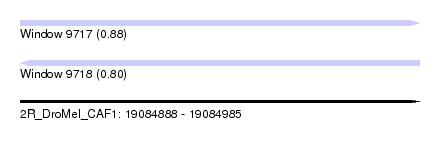
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,084,888 – 19,084,985 |
| Length | 97 |
| Max. P | 0.880227 |

| Location | 19,084,888 – 19,084,985 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -25.72 |
| Energy contribution | -26.17 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
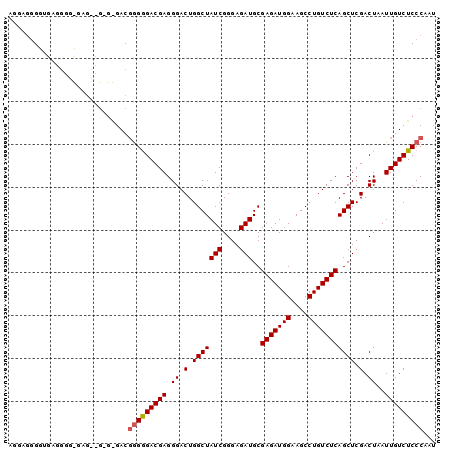
>2R_DroMel_CAF1 19084888 97 + 20766785 AGG---GGUGAGGGGUGAGCGGUGAGACGAGGGACGAGGGACUGGCUAUCGGGAGAUGCGAGAUGGAAGCCUGUCUCAGCUCGACUAAUUGUCUCCCAAU .((---((..(..(((((((..((((((..(((.(......((.((.(((....))))).))......)))))))))))))).)))..)..))))..... ( -33.00) >DroSec_CAF1 27586 82 + 1 AAGAGGG------------------GACGGGAGACGAGGGACUGGCUAUCGGGAGAUGCGAGAUGGAAGCCUGUCUCAGCUCGACUAAUUGUCUCCCAAU ....(..------------------..)(((((((((.((.(.(((((((....)))..(((((((....))))))))))).).))..)))))))))... ( -32.60) >DroSim_CAF1 28106 100 + 1 AGGAGGGGUGAGGGGCGAGUCGAGGGACUGGGGACGAGGGACUGGCUAUCGGGAGAUGCGAGAUGGAAGCCUGUCUCAGCUCGACUAAUUGUCUCCCAAU .....(((.((.((...(((((((....((....)).(((((.(((((((....)))..........)))).)))))..)))))))..)).)).)))... ( -36.70) >consensus AGGAGGGGUGAGGGG_GAG__G_G_GACGGGGGACGAGGGACUGGCUAUCGGGAGAUGCGAGAUGGAAGCCUGUCUCAGCUCGACUAAUUGUCUCCCAAU ............................(((((((((.((.(.(((((((....)))..(((((((....))))))))))).).))..)))))))))... (-25.72 = -26.17 + 0.45)

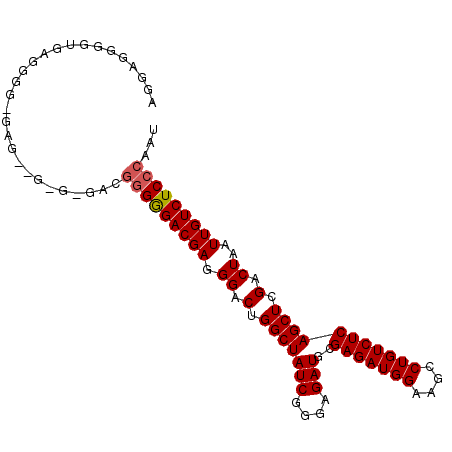
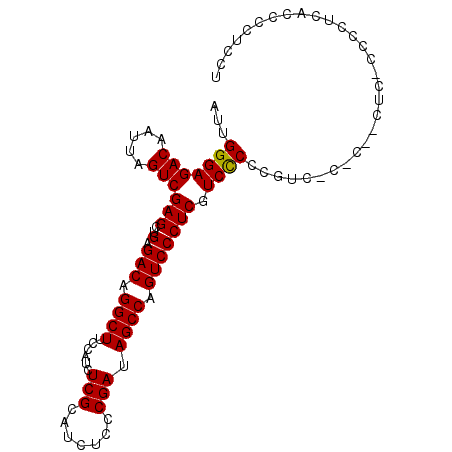
| Location | 19,084,888 – 19,084,985 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -17.15 |
| Energy contribution | -16.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19084888 97 - 20766785 AUUGGGAGACAAUUAGUCGAGCUGAGACAGGCUUCCAUCUCGCAUCUCCCGAUAGCCAGUCCCUCGUCCCUCGUCUCACCGCUCACCCCUCACC---CCU ...(((.(((.....)))(((((((((((((.......((.(((((....))).)).)).........))).))))))..))))..))).....---... ( -27.19) >DroSec_CAF1 27586 82 - 1 AUUGGGAGACAAUUAGUCGAGCUGAGACAGGCUUCCAUCUCGCAUCUCCCGAUAGCCAGUCCCUCGUCUCCCGUC------------------CCCUCUU ...(((((((..((((.....))))(((.((((......(((.......))).)))).)))....)))))))...------------------....... ( -23.90) >DroSim_CAF1 28106 100 - 1 AUUGGGAGACAAUUAGUCGAGCUGAGACAGGCUUCCAUCUCGCAUCUCCCGAUAGCCAGUCCCUCGUCCCCAGUCCCUCGACUCGCCCCUCACCCCUCCU ...(((.((.....((((((((((.((((((.......((.(((((....))).)).))..))).)))..)))...)))))))......)).)))..... ( -23.90) >consensus AUUGGGAGACAAUUAGUCGAGCUGAGACAGGCUUCCAUCUCGCAUCUCCCGAUAGCCAGUCCCUCGUCCCCCGUC_C_C__CUC_CCCCUCACCCCUCCU ...(((((((.....)))(((..(.(((.((((......(((.......))).)))).))))))).)))).............................. (-17.15 = -16.93 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:44 2006