| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,078,191 – 19,078,293 |
| Length | 102 |
| Max. P | 0.895111 |

| Location | 19,078,191 – 19,078,293 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.18 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -20.49 |
| Energy contribution | -21.22 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
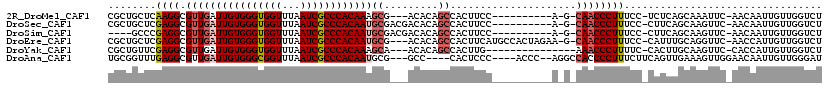
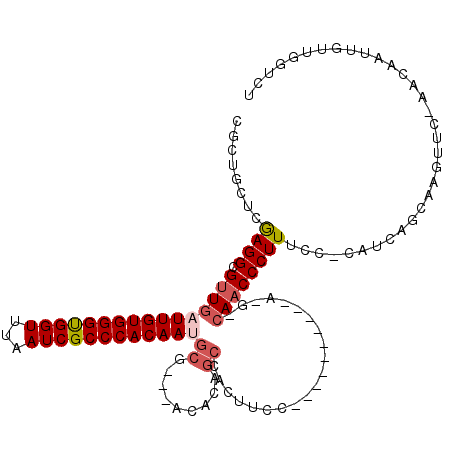
>2R_DroMel_CAF1 19078191 102 - 20766785 CGCUGCUCAAGGCGUUGAUUGUGGGUGGUUUAAUCGCCCACAAAGCG---ACACAGCCACUUCC----------A-G-CAACCCUUUCC-UCUCAGCAAAUUC-AACAAUUGUUGGUCU .((((...((((.((((.(((..(((((((...((((.......)))---)...)))))))..)----------)-)-))))))))...-...))))....((-(((....)))))... ( -34.20) >DroSec_CAF1 20900 105 - 1 CGCUGCUCGAGGCGUUGAUUGUGGGUGGUUUAAUCGCCCACAAUGCGACGACACAGCCACUUCC----------A-G-CAACCCUUUCC-CUUCAGCAAGUUC-AACAAUUGUUGGUCU ...((((.(((((((((((((((((((((...)))))))))))).))))).....((.......----------.-)-).........)-))).))))((..(-(((....))))..)) ( -36.40) >DroSim_CAF1 21507 101 - 1 ----GCCCGAGGCGUUGAUUGUGGGUGGUUUAAUCGCCCACAAUGCGACGACACAGCCACUUCC----------A-G-CAACCCUUUCC-CUUCAGCAAGUUC-AACAAUUGUUGGUCU ----((..(((((((((((((((((((((...)))))))))))).))))).....((.......----------.-)-).........)-)))..)).((..(-(((....))))..)) ( -33.30) >DroEre_CAF1 16972 112 - 1 CGCUGCUCGAGGCGUUGAUUGUGGGUGGUUUAAUCGCCCACAAUGCG---ACACAGCCACUUCAUGCCACUAGAA-G-CAACCCUUUCC-CAUUUGCAGGUUC-AACCAUUGUUGGUCU ..((((....(((((((((((((((((((...)))))))))))).))---))...)))......(((........-)-)).........-.....))))....-.((((....)))).. ( -34.70) >DroYak_CAF1 33986 99 - 1 CGCUGUUCGAGGCGUUGAUUGUGGGUGGUUUAAUCGCCCACAAAGCA---ACACAGCCACUUG---------------AAACCCUUUUC-CACUUGCAAGUUC-CACCAUUGUUGGUCU .(((((.......((((.(((((((((((...)))))))))))..))---))))))).(((((---------------...........-......)))))..-.((((....)))).. ( -27.54) >DroAna_CAF1 16561 106 - 1 UGCGGUUUGAGGCGUUGAUUGUGGGCGGUUUAAUCGCCCACAAUGCG---GCC----CACUCCC----ACCC--AGGCCACCCCUUUCUUCAGUUGAAAGUUGGAACAAUUGUUGGGAU ((.((..((.(((((..((((((((((((...)))))))))))))).---)))----))..)))----)(((--...(((...(((((.......))))).)))(((....)))))).. ( -38.10) >consensus CGCUGCUCGAGGCGUUGAUUGUGGGUGGUUUAAUCGCCCACAAUGCG___ACACAGCCACUUCC__________A_G_CAACCCUUUCC_CAUCAGCAAGUUC_AACAAUUGUUGGUCU ........((((.((((((((((((((((...))))))))))))((.........)).....................))))))))................................. (-20.49 = -21.22 + 0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:42 2006