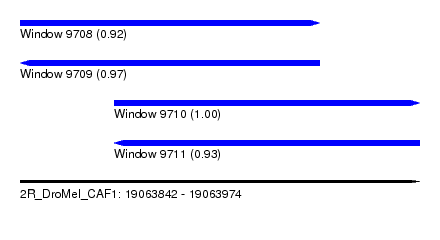
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,063,842 – 19,063,974 |
| Length | 132 |
| Max. P | 0.996167 |

| Location | 19,063,842 – 19,063,941 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.50 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.80 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19063842 99 + 20766785 GUUCGUUGCUGGUUCCGGUUCAAGUUCCAUUUCUUCGCCGCAACUACAGUGCCUAUCGGUUGCGGCGAAUUGCAAUUUUAAAAACCAAUUUUGUUGAAA ....((((((((..(........)..)))....((((((((((((............))))))))))))..)))))....................... ( -25.10) >DroSec_CAF1 6327 99 + 1 GUUCGUUGCUGGUUCCGGUUCCAGUUCCAUUUCUUCGCCGCAACUACAGUUCCUAUCGAUUGCGGCGAAUUGAAAUUUGAAAAACCAAUUUUAUUAAAA .((((..(((((........)))))..)(((((((((((((((................))))))))))..)))))..))).................. ( -24.99) >DroSim_CAF1 6221 99 + 1 GUUCGUUGCUGGUUCCGGUUCCAGUUCCAUUUCUUCGCCGCAACUACAGUUCCUAUCGGUUGCGGCGAAUUGAAAUUUGAAAAACCAAUUUUAUUGAAA .((((..(((((........)))))..)(((((((((((((((((............))))))))))))..)))))..))).................. ( -29.80) >DroEre_CAF1 6274 92 + 1 AUUCGAUGCCGGUUCCGGCUCACGA------UCUUCGCCGCAACUACAGUGUCUAUCGGUUCCGGCGAAUUGGAGUUUUAAUAAC-UAUUUUACUGAAA .(((((.((((....)))))).(((------(..((((((.((((............)))).)))))))))).((((.....)))-)........))). ( -24.10) >DroYak_CAF1 6626 86 + 1 AUUCGGAGCCGGUU------CAAGU------UCUUCGCCGCAAAUACGGUGUCUAUCGGUUGCGGCGAAUUGGAAUUUGAAAAAC-AAUUUUAUUGAAA ..(((....)))((------(((((------((((((((((((...((((....)))).)))))))))...))))))))))....-............. ( -30.50) >consensus GUUCGUUGCUGGUUCCGGUUCAAGUUCCAUUUCUUCGCCGCAACUACAGUGCCUAUCGGUUGCGGCGAAUUGAAAUUUGAAAAACCAAUUUUAUUGAAA .((((..((((....))))..............((((((((((((............)))))))))))).))))......................... (-17.60 = -18.80 + 1.20)


| Location | 19,063,842 – 19,063,941 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.50 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -18.10 |
| Energy contribution | -20.11 |
| Covariance contribution | 2.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19063842 99 - 20766785 UUUCAACAAAAUUGGUUUUUAAAAUUGCAAUUCGCCGCAACCGAUAGGCACUGUAGUUGCGGCGAAGAAAUGGAACUUGAACCGGAACCAGCAACGAAC ...........((((((((...........(((((((((((..((((...)))).))))))))))).....((........))))))))))........ ( -27.40) >DroSec_CAF1 6327 99 - 1 UUUUAAUAAAAUUGGUUUUUCAAAUUUCAAUUCGCCGCAAUCGAUAGGAACUGUAGUUGCGGCGAAGAAAUGGAACUGGAACCGGAACCAGCAACGAAC ...........((((((((((...(((((.(((((((((((..((((...)))).)))))))))))....)))))...)))...)))))))........ ( -28.70) >DroSim_CAF1 6221 99 - 1 UUUCAAUAAAAUUGGUUUUUCAAAUUUCAAUUCGCCGCAACCGAUAGGAACUGUAGUUGCGGCGAAGAAAUGGAACUGGAACCGGAACCAGCAACGAAC ...........((((((((((...(((((.(((((((((((..((((...)))).)))))))))))....)))))...)))...)))))))........ ( -30.20) >DroEre_CAF1 6274 92 - 1 UUUCAGUAAAAUA-GUUAUUAAAACUCCAAUUCGCCGGAACCGAUAGACACUGUAGUUGCGGCGAAGA------UCGUGAGCCGGAACCGGCAUCGAAU .............-................(((((((.(((..((((...)))).))).)))))))..------(((...((((....))))..))).. ( -26.10) >DroYak_CAF1 6626 86 - 1 UUUCAAUAAAAUU-GUUUUUCAAAUUCCAAUUCGCCGCAACCGAUAGACACCGUAUUUGCGGCGAAGA------ACUUG------AACCGGCUCCGAAU .............-....(((((.(((...((((((((((.((........))...))))))))))))------).)))------)).((....))... ( -21.60) >consensus UUUCAAUAAAAUUGGUUUUUCAAAUUCCAAUUCGCCGCAACCGAUAGGCACUGUAGUUGCGGCGAAGAAAUGGAACUUGAACCGGAACCAGCAACGAAC ...........(((((((............(((((((((((..((((...)))).)))))))))))............))))))).............. (-18.10 = -20.11 + 2.01)


| Location | 19,063,873 – 19,063,974 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.91 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -17.35 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19063873 101 + 20766785 UCUUCGCCGCAACUACAGUGCCUAUCGGUUGCGGCGAAUUGCAAUUUUAAAAACCAAUUUUGUUGAAAUUCGAAAGUAAAUGUAACGAAGACUUGCAAAUU-- ..((((((((((((............))))))))))))((((((.((...........(((((((.....(....)......))))))))).))))))...-- ( -28.20) >DroSec_CAF1 6358 103 + 1 UCUUCGCCGCAACUACAGUUCCUAUCGAUUGCGGCGAAUUGAAAUUUGAAAAACCAAUUUUAUUAAAAUUCAAAAGUAAAUAUAACGAAGACCUACAAAAUAG ((((((((((((................)))))))...(((((.(((((.(((.....))).))))).))))).............)))))............ ( -19.79) >DroSim_CAF1 6252 103 + 1 UCUUCGCCGCAACUACAGUUCCUAUCGGUUGCGGCGAAUUGAAAUUUGAAAAACCAAUUUUAUUGAAAUUCAAAAGUAAACAUAACGAAGACCUACAAAAUAG ((((((((((((((............)))))))))(((((.(((.(((......))).))).....)))))...............)))))............ ( -24.40) >DroEre_CAF1 6299 92 + 1 UCUUCGCCGCAACUACAGUGUCUAUCGGUUCCGGCGAAUUGGAGUUUUAAUAAC-UAUUUUACUGAAAUCCC------CUUCAUACGAAAUU----GGAAAUU ..(((((((.((((............)))).)))))))..((..(((((.(((.-....))).)))))..))------.((((........)----))).... ( -17.40) >DroYak_CAF1 6645 102 + 1 UCUUCGCCGCAAAUACGGUGUCUAUCGGUUGCGGCGAAUUGGAAUUUGAAAAAC-AAUUUUAUUGAAAUUCCAAAGUACUUAUUACGAAGAUAUCCAGAAAAA ((((((((((((...((((....)))).)))))))...(((((((((.((.((.-....)).)).)))))))))............)))))............ ( -29.70) >consensus UCUUCGCCGCAACUACAGUGCCUAUCGGUUGCGGCGAAUUGAAAUUUGAAAAACCAAUUUUAUUGAAAUUCAAAAGUAAAUAUAACGAAGACCUACAAAAUAG ((((((((((((((............)))))))))(((((..(((................)))..)))))...............)))))............ (-17.35 = -18.07 + 0.72)

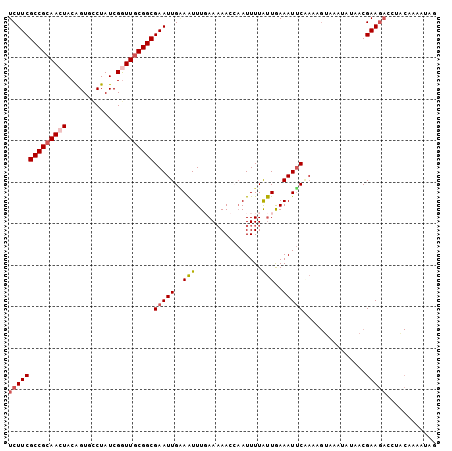
| Location | 19,063,873 – 19,063,974 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.91 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -17.06 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19063873 101 - 20766785 --AAUUUGCAAGUCUUCGUUACAUUUACUUUCGAAUUUCAACAAAAUUGGUUUUUAAAAUUGCAAUUCGCCGCAACCGAUAGGCACUGUAGUUGCGGCGAAGA --...((((((......((.......))(((..((..((((.....))))..))..)))))))))(((((((((((..((((...)))).))))))))))).. ( -26.50) >DroSec_CAF1 6358 103 - 1 CUAUUUUGUAGGUCUUCGUUAUAUUUACUUUUGAAUUUUAAUAAAAUUGGUUUUUCAAAUUUCAAUUCGCCGCAAUCGAUAGGAACUGUAGUUGCGGCGAAGA .......((((((.........)))))).((((((..(((((...)))))...))))))......(((((((((((..((((...)))).))))))))))).. ( -22.20) >DroSim_CAF1 6252 103 - 1 CUAUUUUGUAGGUCUUCGUUAUGUUUACUUUUGAAUUUCAAUAAAAUUGGUUUUUCAAAUUUCAAUUCGCCGCAACCGAUAGGAACUGUAGUUGCGGCGAAGA .......(((((.(........)))))).((((((..(((((...)))))...))))))......(((((((((((..((((...)))).))))))))))).. ( -25.90) >DroEre_CAF1 6299 92 - 1 AAUUUCC----AAUUUCGUAUGAAG------GGGAUUUCAGUAAAAUA-GUUAUUAAAACUCCAAUUCGCCGGAACCGAUAGACACUGUAGUUGCGGCGAAGA .......----.............(------(((.(((.(((((....-.))))).)))))))..(((((((.(((..((((...)))).))).))))))).. ( -22.10) >DroYak_CAF1 6645 102 - 1 UUUUUCUGGAUAUCUUCGUAAUAAGUACUUUGGAAUUUCAAUAAAAUU-GUUUUUCAAAUUCCAAUUCGCCGCAACCGAUAGACACCGUAUUUGCGGCGAAGA ......((((.......(((.....)))(((((((...((((...)))-)..))))))).)))).((((((((((.((........))...)))))))))).. ( -24.40) >consensus CUAUUUUGUAGGUCUUCGUUAUAUUUACUUUGGAAUUUCAAUAAAAUUGGUUUUUCAAAUUCCAAUUCGCCGCAACCGAUAGGCACUGUAGUUGCGGCGAAGA ............(((((...............(((((..(((((((.....))))...)))..)))))((((((((..((((...)))).))))))))))))) (-17.06 = -17.50 + 0.44)

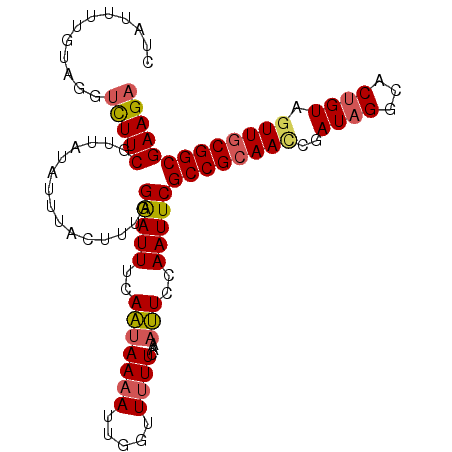

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:37 2006