| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,061,837 – 19,061,948 |
| Length | 111 |
| Max. P | 0.944722 |

| Location | 19,061,837 – 19,061,948 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
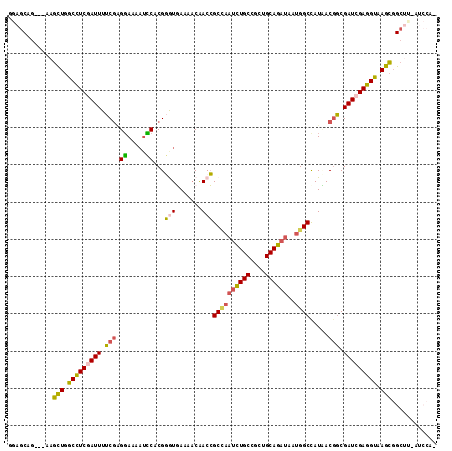
| Mean pairwise identity | 79.79 |
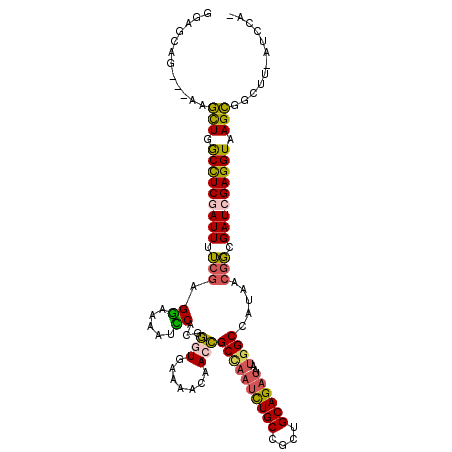
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -29.85 |
| Energy contribution | -29.47 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
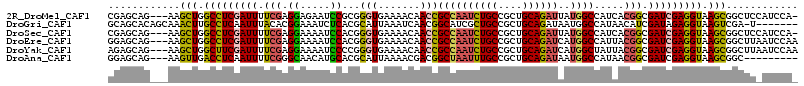
>2R_DroMel_CAF1 19061837 111 - 20766785 CGAGCAG---AAGCUGGCCUCGAUUUUCGAGGAGAAUCCGCGGGUGAAAACAACCGCCAAUCUGCCGCUGCAGAUUAUGGCCAUCACGGCGAUCGAGGUAAGCGGCUCCAUCCA- .((((..---..(((.(((((((((.(((..((....((((((((....))..)))).(((((((....)))))))..))...)).))).))))))))).))).))))......- ( -45.40) >DroGri_CAF1 3783 107 - 1 GCAGCACAGCAAACUUGCCUCAAUUUACACGGAAAUCUCACGCAUUAAAUCAACGGCAUCGCUGCCGCUGCAGAUAAUGGCCAUAACAUCGAUAGAGGUAAGUCGA-U------- ((......))..(((((((((........((((....)).))(((((..(((.(((((....))))).)).)..)))))...............)))))))))...-.------- ( -25.90) >DroSec_CAF1 4352 111 - 1 CGAGCAG---AAGCUGGCCUCGAUUUUCGAGGAAAAUCCACGGGUGAAAACAACCGCCAAUCUGCCGCUGCAGAUUAUGGCCAUCACGGCGAUCGAGGUAAGCGGCUCCAUCCA- .((((..---..(((.(((((((((.(((.((.....))...(((.......)))((((((((((....))))))..)))).....))).))))))))).))).))))......- ( -44.80) >DroEre_CAF1 4208 112 - 1 GGAGCAG---AAGCUGGCCUCGAUUUUCGAGGAAAAUCCACGGGUGAAAACAACCGCCAAUCUGCCGCUGCAGAUCAUGGCCAUUACGGCGAUCGAGGUAAGCGGCUUAAUCCAA (((....---(((((((((((((((.(((.((.....))...(((.......)))((((((((((....))))))..)))).....))).)))))))))...))))))..))).. ( -44.50) >DroYak_CAF1 4536 112 - 1 AGAGCAG---AAGCUGGCUUCGAUUUUCGAGGAAAAUCCCCGGGUGAAAACAACCGCCAAUCUGCCGCUGCAGAUCAUGGCUAUUACGGCGAUCGAGGUAAGCGGCUUAAUCCAA .((((..---..(((.(((((((((.....((.....))((((((.......)))((((((((((....))))))..)))).....))).))))))))).))).))))....... ( -40.00) >DroAna_CAF1 3891 103 - 1 GGAGCAG---AAGUUGACCUCAAUUUUCGGGCAACAUGCACGCAUUAAAACGACGGCUAAUUUGCCGCUGCAGAUAAUGGCCAUAACGGCGAUCGAGGUAAGCGGC--------- ...(((.---..((((.(((........))))))).))).(((........(.((((......)))))(((.(((....(((.....))).)))...))).)))..--------- ( -28.10) >consensus GGAGCAG___AAGCUGGCCUCGAUUUUCGAGGAAAAUCCACGGGUGAAAACAACCGCCAAUCUGCCGCUGCAGAUAAUGGCCAUAACGGCGAUCGAGGUAAGCGGCUU_AUCCA_ ............(((.(((((((((.(((.((.....))...(((.......)))((((((((((....))))))..)))).....))).))))))))).)))............ (-29.85 = -29.47 + -0.38)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:32 2006