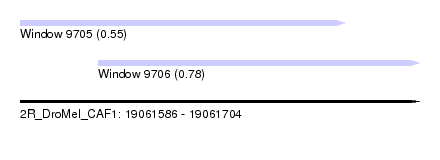
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,061,586 – 19,061,704 |
| Length | 118 |
| Max. P | 0.779271 |

| Location | 19,061,586 – 19,061,682 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 70.38 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -9.13 |
| Energy contribution | -9.60 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
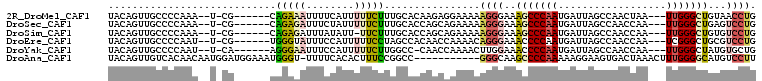
>2R_DroMel_CAF1 19061586 96 + 20766785 UACAGUUGCCCCAAA--U-CG------CAGAAAUUUUCAUUUUUCUUUGCACAAGAGGAAAAAGGGAAAGCCCAAUGAUUAGCCAACUAA---UUGGGCUGUAACCUG ....((((((((...--.-..------..((.....)).(((((((((......))))))))))))..((((((((...(((....))))---))))))))))))... ( -25.50) >DroSec_CAF1 4101 96 + 1 UACAGUUGCCCCAAA--U-CG------CAGAGAUUUCUAUUUUUCUUUGCACCAGCAGAAAAAGGGAAAGCCCAAUGAUUAGCCAACCAA---UUGGGCUGAGUCCUG ...............--.-.(------((((((..........))))))).............((((.((((((((.............)---)))))))...)))). ( -25.72) >DroSim_CAF1 3989 95 + 1 UACAGUUGCCCCAAA--U-CG------CAGAGAUUUAUAUU-UUCUUUGCACCAGCAGAAAAAGGGAAAGCCCAAUGAUUAGCCAACCAA---UUGGGCUGUGUCCUG ...............--.-.(------((((((........-.))))))).............((((.((((((((.............)---)))))))...)))). ( -25.82) >DroEre_CAF1 3943 96 + 1 UACAGUUGCCCCAAU--U-CG------UGGGUAUUUCCAUUUUUCCUAGCCACAACCAAAACAGGGAAACCCCAAUGAUUAGCCAACCAA---UCGGGCUGCGUCCUG ....((.((((....--.-.(------(((.((.............)).))))..........(((....)))...((((........))---)))))).))...... ( -20.02) >DroYak_CAF1 4272 95 + 1 UACAGUUGCCCCAAU--U-CA------AGGGAAUUUCCAUUUUUCUUGGCC-CAACCAAAACUUGGAAACCCCAAUGAUUAGCCAACCAA---UUGGGCUAUGUGCUG ((((...((((.(((--(-..------.(((..((((((......((((..-...))))....)))))).)))..((......))...))---))))))..))))... ( -22.00) >DroAna_CAF1 3638 96 + 1 UACAGUUGUCACAACAAUGGAUGGAAAUGGGU-UUUUCACACUUUCCGGCC-----------GGGCAAGCCCCAAAAAGGAAGUGACUAAACUUUGGGGCAUGUCCUU ....(((((....)))))((.((((((((((.-...)).)).)))))).))-----------(((((.(((((((......(((......)))))))))).))))).. ( -31.40) >consensus UACAGUUGCCCCAAA__U_CG______CAGAAAUUUCCAUUUUUCUUUGCACCAGCAGAAAAAGGGAAACCCCAAUGAUUAGCCAACCAA___UUGGGCUGUGUCCUG ............................(((((........)))))................((((..(((((((..................)))))))...)))). ( -9.13 = -9.60 + 0.48)
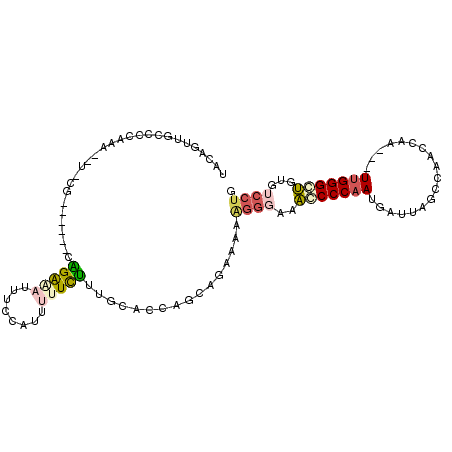
| Location | 19,061,609 – 19,061,704 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 72.20 |
| Mean single sequence MFE | -23.56 |
| Consensus MFE | -11.77 |
| Energy contribution | -12.19 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19061609 95 + 20766785 AUUUUCAUUUUUCUUUGCACAAGAGGAAAAAGGGAAAGCCCAAUGAUUAGCCAACUAA---UUGGGCUGUAACCUGGUCGCACUUUUCAAGGACAUUC ...........((((((...(((.(((...(((...((((((((...(((....))))---)))))))....)))..)).).)))..))))))..... ( -23.40) >DroSec_CAF1 4124 95 + 1 AUUUCUAUUUUUCUUUGCACCAGCAGAAAAAGGGAAAGCCCAAUGAUUAGCCAACCAA---UUGGGCUGAGUCCUGGUCGCACUCUCCAAGGACAUUC ..((((.(((((((..((....))))))))).))))((((((((.............)---)))))))..(((((((.........)).))))).... ( -28.22) >DroSim_CAF1 4012 94 + 1 AUUUAUAUU-UUCUUUGCACCAGCAGAAAAAGGGAAAGCCCAAUGAUUAGCCAACCAA---UUGGGCUGUGUCCUGGUCGCACUCUCCAAGGACAUUC .(((...((-((.(((((....))))).)))).)))((((((((.............)---)))))))(((((((((.........)).))))))).. ( -26.22) >DroEre_CAF1 3966 95 + 1 AUUUCCAUUUUUCCUAGCCACAACCAAAACAGGGAAACCCCAAUGAUUAGCCAACCAA---UCGGGCUGCGUCCUGGUCCCACUUUUCAAGGACGUUC ...............((((............(((....)))..(((((........))---)))))))(((((((..............))))))).. ( -21.04) >DroYak_CAF1 4295 94 + 1 AUUUCCAUUUUUCUUGGCC-CAACCAAAACUUGGAAACCCCAAUGAUUAGCCAACCAA---UUGGGCUAUGUGCUGGUCCCACUUUUCAAGGACGUUC .............((((..-...))))..((((((((.......(((((((((.((..---..))....)).)))))))....))))))))....... ( -17.90) >DroAna_CAF1 3670 84 + 1 -UUUUCACACUUUCCGGCC-----------GGGCAAGCCCCAAAAAGGAAGUGACUAAACUUUGGGGCAUGUCCUUGACCAGUGUUUGAUGGAC--UC -.....(((((....(((.-----------(((((.(((((((......(((......)))))))))).)))))..).))))))).........--.. ( -24.60) >consensus AUUUCCAUUUUUCUUUGCACCAGCAGAAAAAGGGAAACCCCAAUGAUUAGCCAACCAA___UUGGGCUGUGUCCUGGUCCCACUUUUCAAGGACAUUC ..........((((((..............))))))(((((((..................)))))))(((((((((.........))).)))))).. (-11.77 = -12.19 + 0.42)

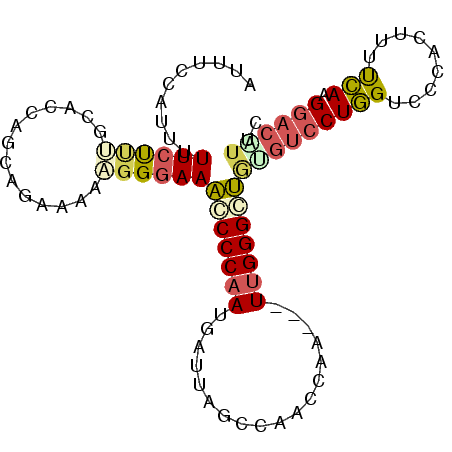
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:30 2006