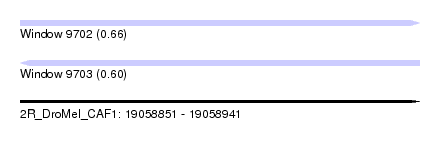
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,058,851 – 19,058,941 |
| Length | 90 |
| Max. P | 0.657203 |

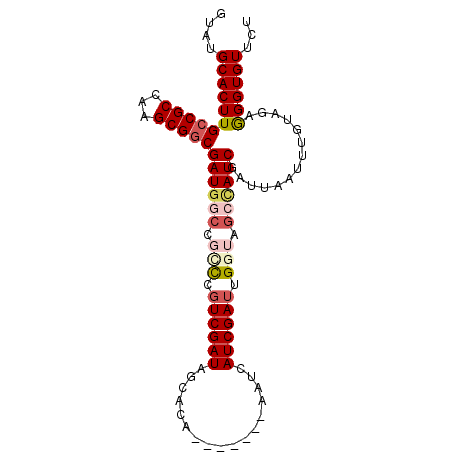
| Location | 19,058,851 – 19,058,941 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 84.09 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.66 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
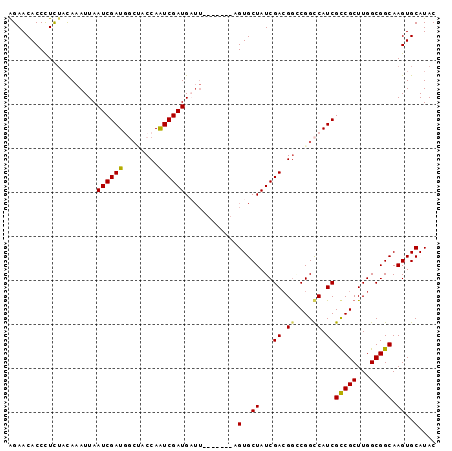
>2R_DroMel_CAF1 19058851 90 + 20766785 AGAACACCCUCUACAAAUUAAUCGAUGGCCACCAAUCGAUGAUU-------AGUGCUAUCGACGAGCGGCCAUCGCCGCUUGGCGGCAAGUGCAUAC ....(((................(((((((.(...((((((...-------.....))))))...).)))))))(((((...)))))..)))..... ( -29.90) >DroSec_CAF1 1315 90 + 1 AGAACACCCUCUACAAAUUAAUCGAUGGCUACCAAUCGAUGAUU-------AGUGCUAUCGACGGGCGGCCAUCGCCGCUUGGCGGCAAGUGCAUAC ....(((................((((((..((..((((((...-------.....))))))..))..))))))(((((...)))))..)))..... ( -33.00) >DroSim_CAF1 1169 90 + 1 AGAACACCCUCUACAAAUUAAUCGAUGGAUACCAAUCGAUGAUU-------UGUGCUAUCGACGGACGGCCAUCGCCGCUUGGCGGCAAGUGCAUAC ....(((.....((((((((.(((((((...)).))))))))))-------)))(((..((((((.((.....))))).)))..)))..)))..... ( -25.20) >DroEre_CAF1 1358 92 + 1 GGAACACCCUCUACACAUUAAUCGAUAUCGCCCAGUCGAUG-----UCAGCCGUGCUAUCGAAGGCCGGUCAUCGCCGCUUGGCGGCAAGUGCAUAC ((.....))....(((.......(((((((......)))))-----)).((((.(((......)))))))....(((((...)))))..)))..... ( -27.30) >DroYak_CAF1 1347 97 + 1 AGAACACCUUUAUUAAAUUAAUCGAUUUCACCCAAUCGAUGUUUAGUCGGCUGUGCUAUCGACGGCCGGCCAUCGUCGCUUGGCGGCAAGUGCAUAC ....(((......(((((..(((((((......))))))))))))(((((((((.......)))))))))....(((((...)))))..)))..... ( -28.10) >consensus AGAACACCCUCUACAAAUUAAUCGAUGGCUACCAAUCGAUGAUU_______AGUGCUAUCGACGGCCGGCCAUCGCCGCUUGGCGGCAAGUGCAUAC ....................((((((........))))))............(..((...((.((....)).))(((((...))))).))..).... (-19.90 = -19.66 + -0.24)

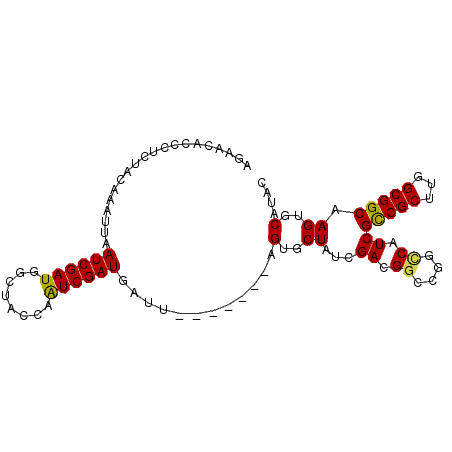
| Location | 19,058,851 – 19,058,941 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 84.09 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -23.43 |
| Energy contribution | -24.91 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19058851 90 - 20766785 GUAUGCACUUGCCGCCAAGCGGCGAUGGCCGCUCGUCGAUAGCACU-------AAUCAUCGAUUGGUGGCCAUCGAUUAAUUUGUAGAGGGUGUUCU ....(((((((((((...)))))((((((((((.((((((......-------....)))))).))))))))))..............))))))... ( -39.00) >DroSec_CAF1 1315 90 - 1 GUAUGCACUUGCCGCCAAGCGGCGAUGGCCGCCCGUCGAUAGCACU-------AAUCAUCGAUUGGUAGCCAUCGAUUAAUUUGUAGAGGGUGUUCU ....(((((((((((...)))))((((((..((.((((((......-------....)))))).))..))))))..............))))))... ( -35.50) >DroSim_CAF1 1169 90 - 1 GUAUGCACUUGCCGCCAAGCGGCGAUGGCCGUCCGUCGAUAGCACA-------AAUCAUCGAUUGGUAUCCAUCGAUUAAUUUGUAGAGGGUGUUCU ....(((((((((((...)))))(((((....)))))......(((-------(((.((((((.((...))))))))..))))))...))))))... ( -27.60) >DroEre_CAF1 1358 92 - 1 GUAUGCACUUGCCGCCAAGCGGCGAUGACCGGCCUUCGAUAGCACGGCUGA-----CAUCGACUGGGCGAUAUCGAUUAAUGUGUAGAGGGUGUUCC ....(((((((((((...)))))((((..(((((...........))))).-----))))..(((.(((.((.....)).))).))).))))))... ( -27.70) >DroYak_CAF1 1347 97 - 1 GUAUGCACUUGCCGCCAAGCGACGAUGGCCGGCCGUCGAUAGCACAGCCGACUAAACAUCGAUUGGGUGAAAUCGAUUAAUUUAAUAAAGGUGUUCU ....(((((((((((((........))).)))).((((((..(((..(((((........).)))))))..))))))...........))))))... ( -25.40) >consensus GUAUGCACUUGCCGCCAAGCGGCGAUGGCCGCCCGUCGAUAGCACA_______AAUCAUCGAUUGGUAGCCAUCGAUUAAUUUGUAGAGGGUGUUCU ....(((((((((((...)))))((((((.(((.((((((.................)))))).))).))))))..............))))))... (-23.43 = -24.91 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:25 2006