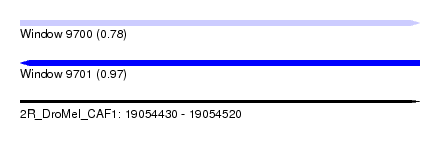
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,054,430 – 19,054,520 |
| Length | 90 |
| Max. P | 0.967123 |

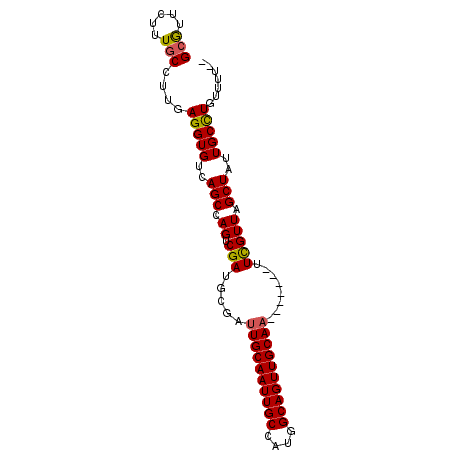
| Location | 19,054,430 – 19,054,520 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 90.86 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -15.57 |
| Energy contribution | -15.82 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19054430 90 + 20766785 --AAAACAGGCAAUAGCUAACGAAUUGCGAAUUGCAACUGCCAUGGCAAUUGCAAUCGCAUCGACUGGCUGACACCUCAAGGCAAAGAAUGC --.....(((...((((((.(((..(((((.((((((.(((....))).))))))))))))))..))))))...)))....(((.....))) ( -28.50) >DroSec_CAF1 1478 83 + 1 --AAAACAGGCAAUAGCUAACGAA-------UUGCAACUGCCAUGGCAAUUGCAAUCGCAUCGACUGGCUGACACCUCAAGGCAAAGAACGC --.....(((...((((((.((((-------((((((.(((....))).)))))))....)))..))))))...)))............... ( -21.00) >DroSim_CAF1 1198 83 + 1 --AAAACAGGCAAUAGCUAACGAA-------UUGCAACUGCCAUGGCAAUUGCAAUCGCAUCGACUGGCUGACACCUCAAGGCAAAGAACGC --.....(((...((((((.((((-------((((((.(((....))).)))))))....)))..))))))...)))............... ( -21.00) >DroEre_CAF1 1611 84 + 1 AAAAAACAAGCAAUAGCUAACAAA--------UGCAACUGCAAUUGCAAUUGCAAUCGCAUCGACUGGCUGACACCUCAAGGCAAAGAACUC .............((((((....(--------(((...((((((....))))))...))))....))))))...((....)).......... ( -15.00) >consensus __AAAACAGGCAAUAGCUAACGAA_______UUGCAACUGCCAUGGCAAUUGCAAUCGCAUCGACUGGCUGACACCUCAAGGCAAAGAACGC .......(((...((((((.............(((((.(((....))).))))).(((...))).))))))...)))............... (-15.57 = -15.82 + 0.25)

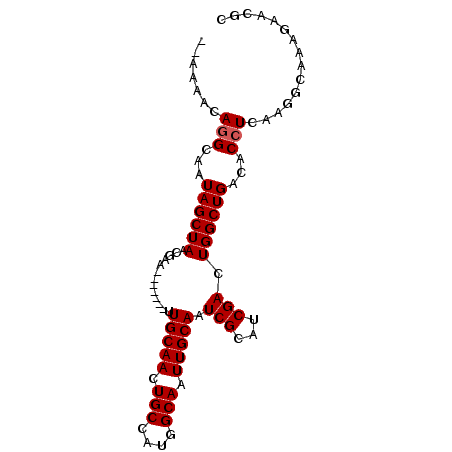
| Location | 19,054,430 – 19,054,520 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 90.86 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -22.96 |
| Energy contribution | -22.89 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19054430 90 - 20766785 GCAUUCUUUGCCUUGAGGUGUCAGCCAGUCGAUGCGAUUGCAAUUGCCAUGGCAGUUGCAAUUCGCAAUUCGUUAGCUAUUGCCUGUUUU-- (((.....)))....(((((..(((.((.((((((((((((((((((....)))))))))).)))))..))))).)))..))))).....-- ( -34.80) >DroSec_CAF1 1478 83 - 1 GCGUUCUUUGCCUUGAGGUGUCAGCCAGUCGAUGCGAUUGCAAUUGCCAUGGCAGUUGCAA-------UUCGUUAGCUAUUGCCUGUUUU-- .........(((....)))..(((.((((.(..((((((((((((((....))))))))))-------.))))...).)))).)))....-- ( -27.40) >DroSim_CAF1 1198 83 - 1 GCGUUCUUUGCCUUGAGGUGUCAGCCAGUCGAUGCGAUUGCAAUUGCCAUGGCAGUUGCAA-------UUCGUUAGCUAUUGCCUGUUUU-- .........(((....)))..(((.((((.(..((((((((((((((....))))))))))-------.))))...).)))).)))....-- ( -27.40) >DroEre_CAF1 1611 84 - 1 GAGUUCUUUGCCUUGAGGUGUCAGCCAGUCGAUGCGAUUGCAAUUGCAAUUGCAGUUGCA--------UUUGUUAGCUAUUGCUUGUUUUUU ((((.....(((....)))...(((.((..((((((((((((((....))))))))))))--------))..)).)))...))))....... ( -24.70) >consensus GCGUUCUUUGCCUUGAGGUGUCAGCCAGUCGAUGCGAUUGCAAUUGCCAUGGCAGUUGCAA_______UUCGUUAGCUAUUGCCUGUUUU__ (((.....)))....(((((..(((.((.(((.....((((((((((....))))))))))........))))).)))..)))))....... (-22.96 = -22.89 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:23 2006