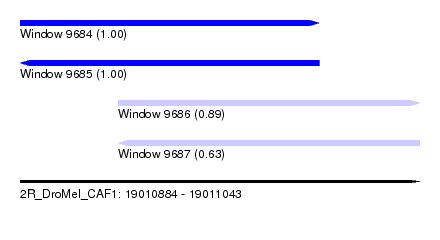
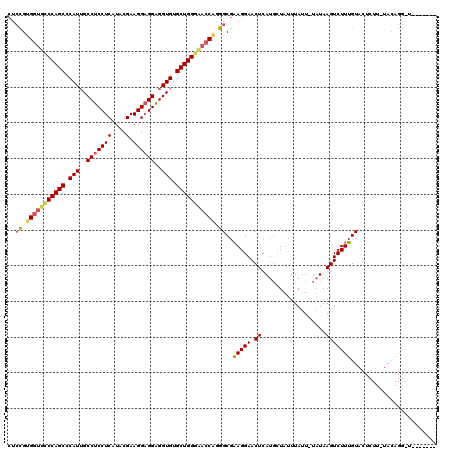
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,010,884 – 19,011,043 |
| Length | 159 |
| Max. P | 0.999681 |

| Location | 19,010,884 – 19,011,003 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.01 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -27.44 |
| Energy contribution | -29.67 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.77 |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19010884 119 + 20766785 CUGUAAAAUCCUGUAAAAGAGUACAAAGACUUAUA-AAUAAAUAGGAUGAGUUCCUUCGCCCGGGUUCCCAGCACACCUCCUCCUUCGUAUGAGGAGGCAAUGGGCUGGGAAACACGGAG .......(((((((....((((......))))...-.....)))))))............(((..((((((((.(((((((((........)))))))...)).))))))))...))).. ( -39.10) >DroSec_CAF1 109144 112 + 1 -------AGCCUGUAGAAGAGUACAAAGACUUAUA-AAUAAAAAGCAUGAGUUCCUUCGCCAUGGUUCCCAGCACACCUCCUCCUUCGUAUGAGGAGGCAAUGGGCUGGACGCCACGGAU -------........((((.(......(((((((.-..........)))))))))))).((.((((..(((((.(((((((((........)))))))...)).)))))..)))).)).. ( -33.90) >DroYak_CAF1 113126 98 + 1 ----------------------ACUAAGACUGAAAGAGUAAAUAACAUGAGUUCCUACGCCCAGGUGUCCAGCACACCCCCGCCUUCGUAUGAGGAGGCCAUGGGCUGGGCACCUCACAG ----------------------.......(((.....(((...(((....)))..)))....(((((((((((...((...((((((......))))))...)))))))))))))..))) ( -33.40) >consensus _______A_CCUGUA_AAGAGUACAAAGACUUAUA_AAUAAAUAGCAUGAGUUCCUUCGCCCAGGUUCCCAGCACACCUCCUCCUUCGUAUGAGGAGGCAAUGGGCUGGGCACCACGGAG ...........................(((((((............)))))))......((.(((((((((((.((...(((((((.....)))))))...)).))))))))))).)).. (-27.44 = -29.67 + 2.23)

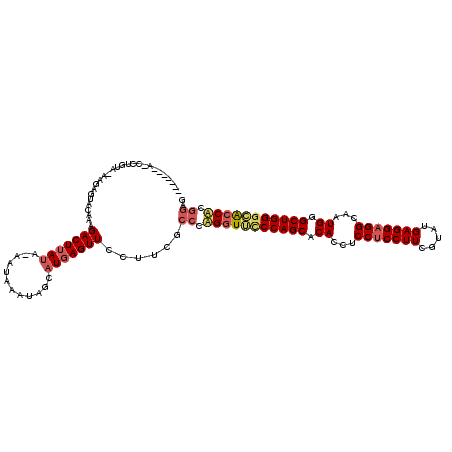
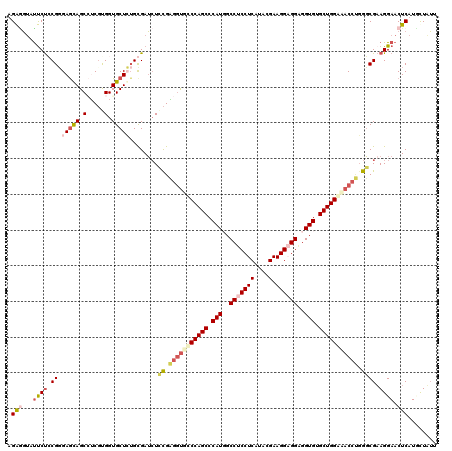
| Location | 19,010,884 – 19,011,003 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.01 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -28.55 |
| Energy contribution | -29.67 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.76 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19010884 119 - 20766785 CUCCGUGUUUCCCAGCCCAUUGCCUCCUCAUACGAAGGAGGAGGUGUGCUGGGAACCCGGGCGAAGGAACUCAUCCUAUUUAUU-UAUAAGUCUUUGUACUCUUUUACAGGAUUUUACAG .((((.(.((((((((.(((..(((((((....).))))))..))).)))))))).)))))...((((.....)))).......-...(((((((.(((......))))))))))..... ( -41.60) >DroSec_CAF1 109144 112 - 1 AUCCGUGGCGUCCAGCCCAUUGCCUCCUCAUACGAAGGAGGAGGUGUGCUGGGAACCAUGGCGAAGGAACUCAUGCUUUUUAUU-UAUAAGUCUUUGUACUCUUCUACAGGCU------- ..((((((..((((((.(((..(((((((....).))))))..))).))))))..)))))).(((((.((....((((......-...))))....)).).))))........------- ( -39.90) >DroYak_CAF1 113126 98 - 1 CUGUGAGGUGCCCAGCCCAUGGCCUCCUCAUACGAAGGCGGGGGUGUGCUGGACACCUGGGCGUAGGAACUCAUGUUAUUUACUCUUUCAGUCUUAGU---------------------- (((.((((.(((((((((...((((.(......).)))).)))((((.....))))))))))((((((((....))).))))))))).))).......---------------------- ( -31.90) >consensus CUCCGUGGUGCCCAGCCCAUUGCCUCCUCAUACGAAGGAGGAGGUGUGCUGGGAACCAGGGCGAAGGAACUCAUGCUAUUUAUU_UAUAAGUCUUUGUACUCUU_UACAGG_U_______ ..((.(((((((((((.(((..(((((((....).))))))..))).))))))))))).))..(((((.((..................)))))))........................ (-28.55 = -29.67 + 1.12)

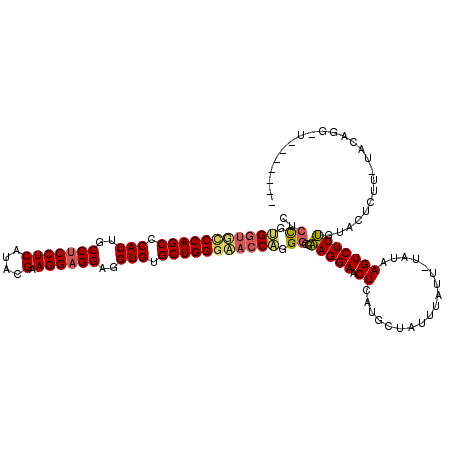
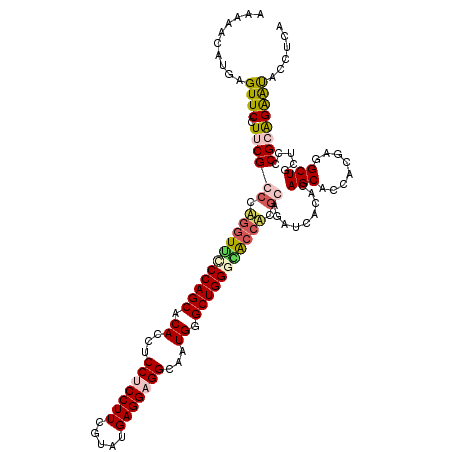
| Location | 19,010,923 – 19,011,043 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.36 |
| Mean single sequence MFE | -42.60 |
| Consensus MFE | -29.26 |
| Energy contribution | -30.95 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19010923 120 + 20766785 AAUAGGAUGAGUUCCUUCGCCCGGGUUCCCAGCACACCUCCUCCUUCGUAUGAGGAGGCAAUGGGCUGGGAAACACGGAGAUCACACAGCACCACGUGGCUGCUCGCGGAGAAUACCUCA .......((((...(((((((((..((((((((.(((((((((........)))))))...)).))))))))...))).........((((((....)).)))).)))))).....)))) ( -48.30) >DroSec_CAF1 109176 120 + 1 AAAAGCAUGAGUUCCUUCGCCAUGGUUCCCAGCACACCUCCUCCUUCGUAUGAGGAGGCAAUGGGCUGGACGCCACGGAUAUCACACAGCACCACGCGGCUACUCCCGGAGAAUUCUUCA ........((((((.((((((.((((..(((((.(((((((((........)))))))...)).)))))..)))).)).........(((........))).....)))))))))).... ( -39.60) >DroEre_CAF1 111263 119 + 1 -CAAAAAUGAGUUCCUUCGACCACGUUUCCAGCACACCACCACCUUCCUAUGAGGAGGCCAUGGGCUGGCGACCUCGCAGCUCGCAGAGCACCGCGAUGCUGUUCCCGCAGGGCAACCCU -.........(((((((((.....(((.(((((...(((...(((((......)))))...)))))))).)))...(((((((((........)))).)))))...)).)))).)))... ( -36.60) >DroYak_CAF1 113144 120 + 1 AAUAACAUGAGUUCCUACGCCCAGGUGUCCAGCACACCCCCGCCUUCGUAUGAGGAGGCCAUGGGCUGGGCACCUCACAGAUCGCUGAGCACCAGGACGCUGCUCCCGCGGAAUACAUCU ......(((.(((((.......(((((((((((...((...((((((......))))))...)))))))))))))........((.(((((.(.....).)))))..))))))).))).. ( -45.90) >consensus AAAAACAUGAGUUCCUUCGCCCAGGUUCCCAGCACACCUCCUCCUUCGUAUGAGGAGGCAAUGGGCUGGGCACCACGCAGAUCACACAGCACCACGAGGCUGCUCCCGCAGAAUACCUCA ..........((((.((((((.(((((((((((.((...(((((((.....)))))))...)).))))))))))).)).........(((........))).....))))))))...... (-29.26 = -30.95 + 1.69)

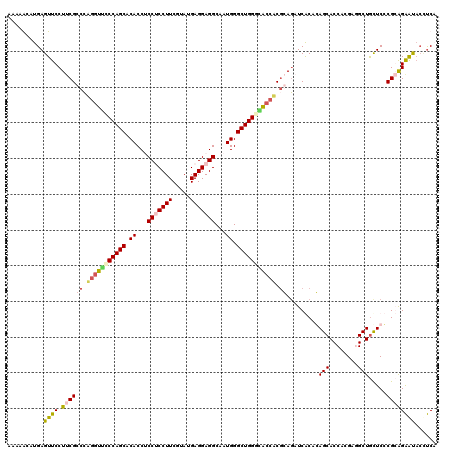
| Location | 19,010,923 – 19,011,043 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.36 |
| Mean single sequence MFE | -49.35 |
| Consensus MFE | -33.06 |
| Energy contribution | -36.12 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19010923 120 - 20766785 UGAGGUAUUCUCCGCGAGCAGCCACGUGGUGCUGUGUGAUCUCCGUGUUUCCCAGCCCAUUGCCUCCUCAUACGAAGGAGGAGGUGUGCUGGGAACCCGGGCGAAGGAACUCAUCCUAUU ((((...((((.(((.((((.((....)))))).........(((.(.((((((((.(((..(((((((....).))))))..))).)))))))).))))))).)))).))))....... ( -51.20) >DroSec_CAF1 109176 120 - 1 UGAAGAAUUCUCCGGGAGUAGCCGCGUGGUGCUGUGUGAUAUCCGUGGCGUCCAGCCCAUUGCCUCCUCAUACGAAGGAGGAGGUGUGCUGGGAACCAUGGCGAAGGAACUCAUGCUUUU (((....(((.(((((....((((((.(((((.....).)))))))))).((((((.(((..(((((((....).))))))..))).))))))..)).))).))).....)))....... ( -45.30) >DroEre_CAF1 111263 119 - 1 AGGGUUGCCCUGCGGGAACAGCAUCGCGGUGCUCUGCGAGCUGCGAGGUCGCCAGCCCAUGGCCUCCUCAUAGGAAGGUGGUGGUGUGCUGGAAACGUGGUCGAAGGAACUCAUUUUUG- .(((((..(((.(((...((((.((((((....))))))))))(.(.((..(((((((((.((((((.....)).)))).))))...)))))..)).).)))).)))))))).......- ( -49.30) >DroYak_CAF1 113144 120 - 1 AGAUGUAUUCCGCGGGAGCAGCGUCCUGGUGCUCAGCGAUCUGUGAGGUGCCCAGCCCAUGGCCUCCUCAUACGAAGGCGGGGGUGUGCUGGACACCUGGGCGUAGGAACUCAUGUUAUU ..(((..((((((....)).((((((.((((..(((((((((((((((.(.(((.....))).).)))))).((....)).)))).)))))..)))).)))))).))))..)))...... ( -51.60) >consensus AGAGGUAUUCUCCGGGAGCAGCCUCGUGGUGCUCUGCGAUCUCCGAGGUGCCCAGCCCAUGGCCUCCUCAUACGAAGGAGGAGGUGUGCUGGAAACCUGGGCGAAGGAACUCAUGCUAUU .(((...((((.((.(((((.(.....).)))))........((.(((((((((((.(((..(((((((....).))))))..))).))))))))))).)))).)))).)))........ (-33.06 = -36.12 + 3.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:08 2006