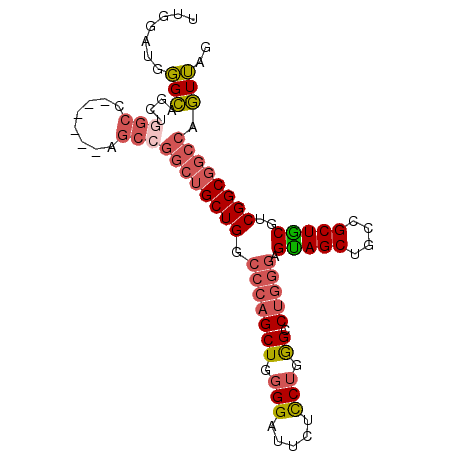
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,998,139 – 18,998,229 |
| Length | 90 |
| Max. P | 0.784325 |

| Location | 18,998,139 – 18,998,229 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 83.19 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -26.04 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18998139 90 + 20766785 CUAACUGGCCGCCGACGCAGCGGCAGCUACUCCCAGGCUCAGGAGAAUCCCCAGCUGGGCCAGCAGCCGGCU------GGACAGUUGCCCAUCCAA .(((((((((((.......))))).....((((........)))).....((((((((........))))))------)).))))))......... ( -34.20) >DroPse_CAF1 100454 90 + 1 CUUAGUGGCCGCCGGCGCAGCGGCAGCUGCACCCAGGCCCAGA----CCCACAGC--GUCCAGCCGAGCGCCGCCACGGUCCAGAGCCAGGUCCAG ....(((((((((((((((((....)))))...........((----(.......--)))..)))).)))..)))))((.((.......)).)).. ( -36.90) >DroSec_CAF1 96463 90 + 1 CUAACGGGCCGCCGACGCAGCGGCAGCUACUCUCAGGCCCAGGAGAAUCCCCAGCUGGGCCAGCAGCCGGCU------GGCCAACUGCCCAUCCAA .....(((((((((......))))((.....))..))))).(((.......(((...(((((((.....)))------))))..)))....))).. ( -36.80) >DroSim_CAF1 101066 90 + 1 CUAACGGGCCGCCGACGCAGCGGCAGCUACUCCCAGGCCCAGGAGAAUCCCCAGCUGGGCCAGCAGCCGGCU------GGCCAACUGCCCAUCCAA .....(((((((((......)))).(.......).))))).(((.......(((...(((((((.....)))------))))..)))....))).. ( -36.60) >DroEre_CAF1 98978 90 + 1 CUAACUGGACGCCGCCGCAGCGGCAGCUACUCCCAGGCCCAGGAGAACCCCCAGCUGGGCCAGCAGCCGGCU------GGCCAGCUGCCCAUCCAA .....((((.(((((....))))).....((((........))))......(((((((.(((((.....)))------)))))))))....)))). ( -40.30) >DroYak_CAF1 100427 90 + 1 CUAACUGGCCGCCGCCGUAGCGGCAGCUACUCCCAGGCCCAGGAGAAUCCCCAGCUGGGCCAGCAGCCCGCC------GGCCAGUUGCCCAUCCAA .((((((((((((((....))))).(((.((....(((((((..(......)..))))))))).))).....------)))))))))......... ( -41.70) >consensus CUAACUGGCCGCCGACGCAGCGGCAGCUACUCCCAGGCCCAGGAGAAUCCCCAGCUGGGCCAGCAGCCGGCU______GGCCAGCUGCCCAUCCAA ......(((.((((......)))).))).......(((((((............))))))).((((..((((......))))..))))........ (-26.04 = -26.90 + 0.86)

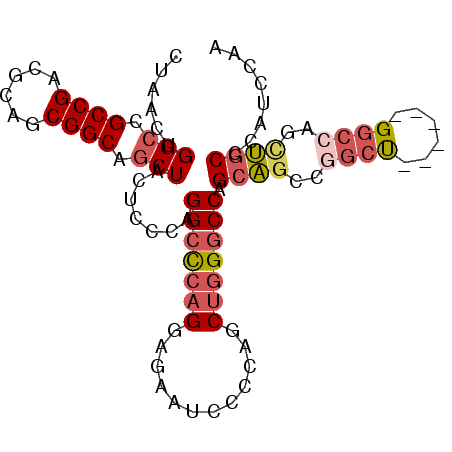

| Location | 18,998,139 – 18,998,229 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 83.19 |
| Mean single sequence MFE | -51.58 |
| Consensus MFE | -31.11 |
| Energy contribution | -32.17 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18998139 90 - 20766785 UUGGAUGGGCAACUGUCC------AGCCGGCUGCUGGCCCAGCUGGGGAUUCUCCUGAGCCUGGGAGUAGCUGCCGCUGCGUCGGCGGCCAGUUAG ..((((((....))))))------(((.((((((((((((((((.(((.....))).)).))))..(((((....))))))))))))))).))).. ( -50.50) >DroPse_CAF1 100454 90 - 1 CUGGACCUGGCUCUGGACCGUGGCGGCGCUCGGCUGGAC--GCUGUGGG----UCUGGGCCUGGGUGCAGCUGCCGCUGCGCCGGCGGCCACUAAG ..(((((((((((..(.(((.(((...)))))))..)).--)))..)))----))).(((((.((((((((....)))))))).).))))...... ( -46.90) >DroSec_CAF1 96463 90 - 1 UUGGAUGGGCAGUUGGCC------AGCCGGCUGCUGGCCCAGCUGGGGAUUCUCCUGGGCCUGAGAGUAGCUGCCGCUGCGUCGGCGGCCCGUUAG ...(((((((.(((((((------((((((((((((((((((..(((....))))))))))....))))))))..)))).)))))).))))))).. ( -51.40) >DroSim_CAF1 101066 90 - 1 UUGGAUGGGCAGUUGGCC------AGCCGGCUGCUGGCCCAGCUGGGGAUUCUCCUGGGCCUGGGAGUAGCUGCCGCUGCGUCGGCGGCCCGUUAG ...(((((((.(((((((------(((.(((.((((.(((((((.(((.....))).)).)))))..)))).))))))).)))))).))))))).. ( -54.40) >DroEre_CAF1 98978 90 - 1 UUGGAUGGGCAGCUGGCC------AGCCGGCUGCUGGCCCAGCUGGGGGUUCUCCUGGGCCUGGGAGUAGCUGCCGCUGCGGCGGCGUCCAGUUAG ((((((((((((((((..------..))))))))).(((((((.((.(((((((((......))))).)))).)))))).)))..))))))).... ( -52.90) >DroYak_CAF1 100427 90 - 1 UUGGAUGGGCAACUGGCC------GGCGGGCUGCUGGCCCAGCUGGGGAUUCUCCUGGGCCUGGGAGUAGCUGCCGCUACGGCGGCGGCCAGUUAG ..........((((((((------....(((((((..(((((((.(((.....))).)).))))))))))))(((((....))))))))))))).. ( -53.40) >consensus UUGGAUGGGCAGCUGGCC______AGCCGGCUGCUGGCCCAGCUGGGGAUUCUCCUGGGCCUGGGAGUAGCUGCCGCUGCGUCGGCGGCCAGUUAG .......(((....(((........)))((((((((.(((((((.(((.....))).)).))))).(((((....)))))..)))))))).))).. (-31.11 = -32.17 + 1.06)

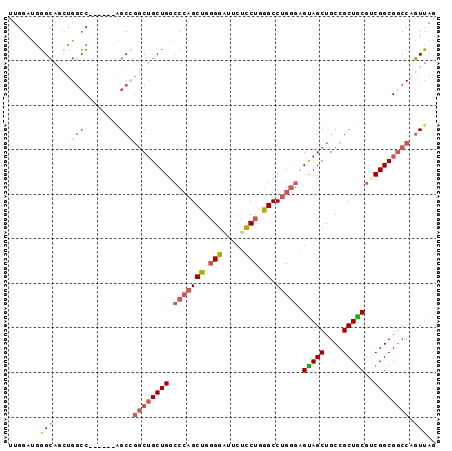
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:02 2006