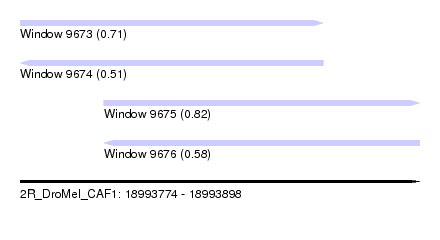
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,993,774 – 18,993,898 |
| Length | 124 |
| Max. P | 0.818344 |

| Location | 18,993,774 – 18,993,868 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.77 |
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -12.39 |
| Energy contribution | -13.01 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18993774 94 + 20766785 AGUGCAACAGUGCUCUUUUUUGUGC-C-------ACACACGCA--CAGCAAUUAUCGCUGCUUCUC--------GCGUUCAUCAGGCACUUUGGCAGCAGCCAGCUGCUAAG ((..(....)..)).......((((-(-------....((((.--.((((........))))....--------))))......))))).((((((((.....)))))))). ( -28.70) >DroSec_CAF1 91928 86 + 1 ----------UGCUCCUUUUUGUGC-C-------ACACACGCACACAGCAAUUAUCGCUGCUUCUC--------GCGUUCAUCACGCACUUUGGCAGCAGCCAGCUGCUAAG ----------..........(((((-.-------......)))))((((.......))))......--------((((.....))))...((((((((.....)))))))). ( -25.00) >DroSim_CAF1 96846 86 + 1 ----------UGCUCUUUUUUGUGC-C-------ACACACGCACACAGCAAUUAUCGCUGCUUCUC--------GCGUUCAUCACGCACUUUGGCAGCAGCCAGCUGCUAAG ----------..........(((((-.-------......)))))((((.......))))......--------((((.....))))...((((((((.....)))))))). ( -25.00) >DroEre_CAF1 94754 84 + 1 ----------UGCUCUUUUUUGUGC-C-------ACACACGCA--CAGCAAUUAUCGCCGCUUCUC--------GCGUUCAUCACGCACUUUGGCAGCAGCCGGCUGCCAAG ----------.((.......(((((-.-------......)))--))((.......)).)).....--------((((.....))))...((((((((.....)))))))). ( -24.90) >DroWil_CAF1 125269 100 + 1 ----------AAUUCGUUUGUGUGUGUAUAGAGAAUAGAGUCG--AAACAAUUAUCGCUGAUUCGCCGACUGCUCUCGCAGUCACGCACUUUGACUGCAGCCAGAUGCUGAA ----------.....(((...((((((.....((((..((.((--(........))))).))))...((((((....))))))))))))...)))..((((.....)))).. ( -29.30) >DroYak_CAF1 95154 84 + 1 ----------UGCUCUUUUUUGUGC-C-------ACACACGCA--CAGCAAUUAUCGCCGCUUCUC--------GCGUUCAUCACGCACUUUGGCAGCAGCCAGCUGCUAAG ----------.((.......(((((-.-------......)))--))((.......)).)).....--------((((.....))))...((((((((.....)))))))). ( -22.70) >consensus __________UGCUCUUUUUUGUGC_C_______ACACACGCA__CAGCAAUUAUCGCUGCUUCUC________GCGUUCAUCACGCACUUUGGCAGCAGCCAGCUGCUAAG .....................((((...............((.....))......(((................)))........)))).((((((((.....)))))))). (-12.39 = -13.01 + 0.61)


| Location | 18,993,774 – 18,993,868 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.77 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -13.78 |
| Energy contribution | -13.01 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.47 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18993774 94 - 20766785 CUUAGCAGCUGGCUGCUGCCAAAGUGCCUGAUGAACGC--------GAGAAGCAGCGAUAAUUGCUG--UGCGUGUGU-------G-GCACAAAAAAGAGCACUGUUGCACU ....(((((((((....))))..(((((......((((--------(....(((((((...))))))--).)))))..-------)-)))).............)))))... ( -35.70) >DroSec_CAF1 91928 86 - 1 CUUAGCAGCUGGCUGCUGCCAAAGUGCGUGAUGAACGC--------GAGAAGCAGCGAUAAUUGCUGUGUGCGUGUGU-------G-GCACAAAAAGGAGCA---------- ..((((((....))))))((....((.((.(((.((((--------(....(((((((...))))))).))))).)))-------.-)).))....))....---------- ( -28.70) >DroSim_CAF1 96846 86 - 1 CUUAGCAGCUGGCUGCUGCCAAAGUGCGUGAUGAACGC--------GAGAAGCAGCGAUAAUUGCUGUGUGCGUGUGU-------G-GCACAAAAAAGAGCA---------- ....((.((((((....))))....))(((....((((--------(....(((((((...)))))))...)))))..-------.-.)))........)).---------- ( -28.40) >DroEre_CAF1 94754 84 - 1 CUUGGCAGCCGGCUGCUGCCAAAGUGCGUGAUGAACGC--------GAGAAGCGGCGAUAAUUGCUG--UGCGUGUGU-------G-GCACAAAAAAGAGCA---------- .((((((((.....)))))))).....(((....((((--------(....(((((((...))))))--).)))))..-------.-.)))...........---------- ( -31.00) >DroWil_CAF1 125269 100 - 1 UUCAGCAUCUGGCUGCAGUCAAAGUGCGUGACUGCGAGAGCAGUCGGCGAAUCAGCGAUAAUUGUUU--CGACUCUAUUCUCUAUACACACACAAACGAAUU---------- ..((((.....))))........(((.((((((((....))))))((.((((.(((((........)--)).))..)))).))....)).))).........---------- ( -26.40) >DroYak_CAF1 95154 84 - 1 CUUAGCAGCUGGCUGCUGCCAAAGUGCGUGAUGAACGC--------GAGAAGCGGCGAUAAUUGCUG--UGCGUGUGU-------G-GCACAAAAAAGAGCA---------- ....((.((((((....))))....))(((....((((--------(....(((((((...))))))--).)))))..-------.-.)))........)).---------- ( -27.10) >consensus CUUAGCAGCUGGCUGCUGCCAAAGUGCGUGAUGAACGC________GAGAAGCAGCGAUAAUUGCUG__UGCGUGUGU_______G_GCACAAAAAAGAGCA__________ (((.(((((.....)))))....((((.......((((..............((((((...)))))).....))))...........))))......)))............ (-13.78 = -13.01 + -0.77)



| Location | 18,993,800 – 18,993,898 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.98 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -20.59 |
| Energy contribution | -20.65 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18993800 98 + 20766785 ACACACGCA--CAGCAAUUAUCGCUGCUUCUC--------GCGUUCAUCAGGCACUUUGGCAGCAGCCAGCUGCUAAGUAACUUCAUUUGACUUGGCUACACGCCAAA------ ....((((.--.((((........))))....--------))))(((..((..((((.((((((.....))))))))))..)).....))).(((((.....))))).------ ( -26.30) >DroSec_CAF1 91944 100 + 1 ACACACGCACACAGCAAUUAUCGCUGCUUCUC--------GCGUUCAUCACGCACUUUGGCAGCAGCCAGCUGCUAAGUAACUUCAUUUGACUUGGCUACACGCCAAA------ ...........((((.......))))......--------((((.....))))((((.((((((.....)))))))))).............(((((.....))))).------ ( -26.40) >DroSim_CAF1 96862 100 + 1 ACACACGCACACAGCAAUUAUCGCUGCUUCUC--------GCGUUCAUCACGCACUUUGGCAGCAGCCAGCUGCUAAGUAACUUCAUUUGACUUGGCUACACGCCAAA------ ...........((((.......))))......--------((((.....))))((((.((((((.....)))))))))).............(((((.....))))).------ ( -26.40) >DroEre_CAF1 94770 104 + 1 ACACACGCA--CAGCAAUUAUCGCCGCUUCUC--------GCGUUCAUCACGCACUUUGGCAGCAGCCGGCUGCCAAGUAACUUCAUUUGACUUGGCUGCACGCCAAAGCCAAA ......((.--..((.......)).)).....--------((((.....)))).(((((((.(((((((...(.(((((......))))).).)))))))..)))))))..... ( -29.60) >DroYak_CAF1 95170 98 + 1 ACACACGCA--CAGCAAUUAUCGCCGCUUCUC--------GCGUUCAUCACGCACUUUGGCAGCAGCCAGCUGCUAAGUAACUUCAUUUGACUUGGCUACACGCCAAA------ ......((.--..((.......)).)).....--------((((.....))))((((.((((((.....)))))))))).............(((((.....))))).------ ( -23.50) >DroPer_CAF1 96252 112 + 1 AGAUAUAGA--CAACAAUUAUCUUUGAUUCUCGCUCUGCUGCGUCAGUCACGCACUUUGACGGCACAGAGAUGCUAAAUAACUUAAUUUUCUGUUGCUGCAUGCUGCAUAGAGG (((((....--.......))))).(((((..(((......)))..))))).(((...((.(((((((((((...(((.....)))..)))))).)))))))...)))....... ( -25.50) >consensus ACACACGCA__CAGCAAUUAUCGCUGCUUCUC________GCGUUCAUCACGCACUUUGGCAGCAGCCAGCUGCUAAGUAACUUCAUUUGACUUGGCUACACGCCAAA______ ...........((((.......))))..............((((.....))))..(((((((((.....)))))))))..............(((((.....)))))....... (-20.59 = -20.65 + 0.06)

| Location | 18,993,800 – 18,993,898 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.98 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -21.29 |
| Energy contribution | -21.85 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18993800 98 - 20766785 ------UUUGGCGUGUAGCCAAGUCAAAUGAAGUUACUUAGCAGCUGGCUGCUGCCAAAGUGCCUGAUGAACGC--------GAGAAGCAGCGAUAAUUGCUG--UGCGUGUGU ------(((((((.(((((((.((....(((......)))...))))))))))))))))...........((((--------(....(((((((...))))))--).))))).. ( -34.00) >DroSec_CAF1 91944 100 - 1 ------UUUGGCGUGUAGCCAAGUCAAAUGAAGUUACUUAGCAGCUGGCUGCUGCCAAAGUGCGUGAUGAACGC--------GAGAAGCAGCGAUAAUUGCUGUGUGCGUGUGU ------(((((((.(((((((.((....(((......)))...))))))))))))))))...........((((--------(....(((((((...)))))))...))))).. ( -34.60) >DroSim_CAF1 96862 100 - 1 ------UUUGGCGUGUAGCCAAGUCAAAUGAAGUUACUUAGCAGCUGGCUGCUGCCAAAGUGCGUGAUGAACGC--------GAGAAGCAGCGAUAAUUGCUGUGUGCGUGUGU ------(((((((.(((((((.((....(((......)))...))))))))))))))))...........((((--------(....(((((((...)))))))...))))).. ( -34.60) >DroEre_CAF1 94770 104 - 1 UUUGGCUUUGGCGUGCAGCCAAGUCAAAUGAAGUUACUUGGCAGCCGGCUGCUGCCAAAGUGCGUGAUGAACGC--------GAGAAGCGGCGAUAAUUGCUG--UGCGUGUGU ....(((((((((.((((((..(((((..........)))))....))))))))))))))).........((((--------(....(((((((...))))))--).))))).. ( -40.90) >DroYak_CAF1 95170 98 - 1 ------UUUGGCGUGUAGCCAAGUCAAAUGAAGUUACUUAGCAGCUGGCUGCUGCCAAAGUGCGUGAUGAACGC--------GAGAAGCGGCGAUAAUUGCUG--UGCGUGUGU ------(((((((.(((((((.((....(((......)))...))))))))))))))))...........((((--------(....(((((((...))))))--).))))).. ( -33.30) >DroPer_CAF1 96252 112 - 1 CCUCUAUGCAGCAUGCAGCAACAGAAAAUUAAGUUAUUUAGCAUCUCUGUGCCGUCAAAGUGCGUGACUGACGCAGCAGAGCGAGAAUCAAAGAUAAUUGUUG--UCUAUAUCU ......(((.....)))(((((((.((((......)))).((.(((((((((.((((.......)))).).))))).)))))...............))))))--)........ ( -22.50) >consensus ______UUUGGCGUGUAGCCAAGUCAAAUGAAGUUACUUAGCAGCUGGCUGCUGCCAAAGUGCGUGAUGAACGC________GAGAAGCAGCGAUAAUUGCUG__UGCGUGUGU ......(((((((.(((((((..((....)).(((....)))...))))))))))))))..((((.....))))..............((((((...))))))........... (-21.29 = -21.85 + 0.56)

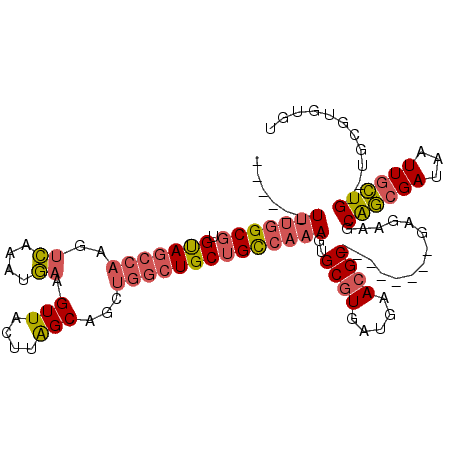

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:57 2006