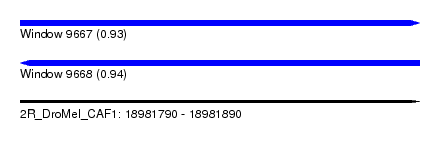
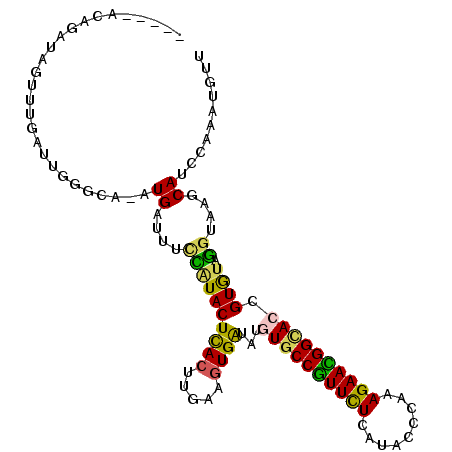
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,981,790 – 18,981,890 |
| Length | 100 |
| Max. P | 0.943169 |

| Location | 18,981,790 – 18,981,890 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 74.03 |
| Mean single sequence MFE | -24.89 |
| Consensus MFE | -16.57 |
| Energy contribution | -17.59 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18981790 100 + 20766785 AACAUUUGGAUGCUUACCUACACGGUGCCGUUCUUUGGUUAUGAAAACGGCACAUAUCACUUCAAGUGAGUAUGGAAAUCAU-UGCCCACUGG--GGGUCUGA----- ........((((.((.(((((...((((((((.(((......)))))))))))...((((.....))))))).)).)).)))-)((((.....--))))....----- ( -27.10) >DroVir_CAF1 111953 93 + 1 AACAUUUGGAUGCUAACGUACACGGUGCCAUUCUUUGGCUAUGAGAAUGGCAGCUAUCACUUCAAGUAAGUAACA---GCAGAAAA-GUCAAAAACA----------- ....(((((.((((....(((..(.((((((((((.......)))))))))).)....((.....))..)))..)---))).....-.)))))....----------- ( -22.10) >DroSec_CAF1 80017 102 + 1 AACAUUUGGAUGCUUACCUACACGGUACCGUUCUUCGGGUAUGAGAACGGCACAUAUCACUUCAAGUGAGUACGGAAAUCAA-UGCCCAAUCAAACUAUCUGU----- .......(((((..((((.....)))).)))))...((((((((...((..((...((((.....)))))).))....)).)-)))))...............----- ( -21.40) >DroSim_CAF1 84653 102 + 1 AACAUCUGGAUGCUUACCUACACGGUGCCGUUCUUUGGGUAUGAGAACGGCACAUAUCACUUCAAGUGAGUAUGGAUAUCAA-UAUCCAAUCAAAGUAUCUGU----- .......((((((((...(((...(((((((((((.......)))))))))))...((((.....)))))))((((((....-))))))....))))))))..----- ( -37.20) >DroWil_CAF1 107014 91 + 1 AAUAUAUGGAUGCUAACCUAUACGGUUCCAUUCUUUGGCUUUGAGAACGGCAGCUAUCAUUUCAAGUAAGUACA--AACCAG-AAU-GAAACAAA------------- .......(((((..((((.....)))).)))))..((((((((....))).)))))((((((...((.......--.))..)-)))-))......------------- ( -12.70) >DroYak_CAF1 83203 107 + 1 AACAUUUGGAUGCUCACCUACACGGUGCCGUUCUUUGGGUAUGAGAACGGCACUUACCACUUCAAGUGAGUAUGGAAAUCAU-GGCUUAAUCAAACUAUUGGUAUAUU ...((((..((((((((......((((((((((((.......))))))))))))...........))))))))..))))...-......(((((....)))))..... ( -28.83) >consensus AACAUUUGGAUGCUUACCUACACGGUGCCGUUCUUUGGGUAUGAGAACGGCACAUAUCACUUCAAGUGAGUAUGGAAAUCAA_UACCCAAUCAAACUAUCUGU_____ ..........(((((((.......(((((((((((.......)))))))))))............))))))).................................... (-16.57 = -17.59 + 1.03)

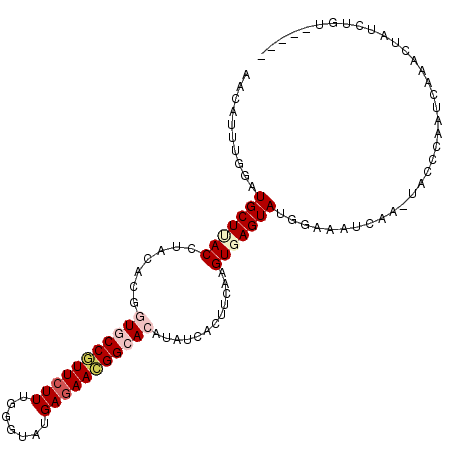
| Location | 18,981,790 – 18,981,890 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 74.03 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -16.37 |
| Energy contribution | -15.60 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18981790 100 - 20766785 -----UCAGACCC--CCAGUGGGCA-AUGAUUUCCAUACUCACUUGAAGUGAUAUGUGCCGUUUUCAUAACCAAAGAACGGCACCGUGUAGGUAAGCAUCCAAAUGUU -----(((...((--(....)))..-.)))...((((((((((.....))))...((((((((((.........)))))))))).)))).))..(((((....))))) ( -25.30) >DroVir_CAF1 111953 93 - 1 -----------UGUUUUUGAC-UUUUCUGC---UGUUACUUACUUGAAGUGAUAGCUGCCAUUCUCAUAGCCAAAGAAUGGCACCGUGUACGUUAGCAUCCAAAUGUU -----------((((..((((-......((---((((((((.....))))))))))(((((((((.........)))))))))....)).))..)))).......... ( -26.20) >DroSec_CAF1 80017 102 - 1 -----ACAGAUAGUUUGAUUGGGCA-UUGAUUUCCGUACUCACUUGAAGUGAUAUGUGCCGUUCUCAUACCCGAAGAACGGUACCGUGUAGGUAAGCAUCCAAAUGUU -----...(((.(((((.(((.((.-.........(((.((((.....)))))))(((((((((((......).)))))))))).)).))).))))))))........ ( -25.80) >DroSim_CAF1 84653 102 - 1 -----ACAGAUACUUUGAUUGGAUA-UUGAUAUCCAUACUCACUUGAAGUGAUAUGUGCCGUUCUCAUACCCAAAGAACGGCACCGUGUAGGUAAGCAUCCAGAUGUU -----((..((((......((((((-....))))))...((((.....))))...((((((((((.........)))))))))).))))..)).(((((....))))) ( -29.00) >DroWil_CAF1 107014 91 - 1 -------------UUUGUUUC-AUU-CUGGUU--UGUACUUACUUGAAAUGAUAGCUGCCGUUCUCAAAGCCAAAGAAUGGAACCGUAUAGGUUAGCAUCCAUAUAUU -------------.(..((((-(..-..(((.--...)))....)))))..)..(((.(((((((.........)))))))((((.....)))))))........... ( -19.70) >DroYak_CAF1 83203 107 - 1 AAUAUACCAAUAGUUUGAUUAAGCC-AUGAUUUCCAUACUCACUUGAAGUGGUAAGUGCCGUUCUCAUACCCAAAGAACGGCACCGUGUAGGUGAGCAUCCAAAUGUU ....((((....((((....))))(-(((.......(((.(((.....)))))).((((((((((.........))))))))))))))..))))(((((....))))) ( -27.20) >consensus _____ACAGAUAGUUUGAUUGGGCA_AUGAUUUCCAUACUCACUUGAAGUGAUAUGUGCCGUUCUCAUACCCAAAGAACGGCACCGUGUAGGUAAGCAUCCAAAUGUU ...........................((....((((((((((.....))))...((((((((((.........)))))))))).)))).))....)).......... (-16.37 = -15.60 + -0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:49 2006