
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,081,090 – 3,081,193 |
| Length | 103 |
| Max. P | 0.759576 |

| Location | 3,081,090 – 3,081,193 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 74.76 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -18.60 |
| Energy contribution | -17.63 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
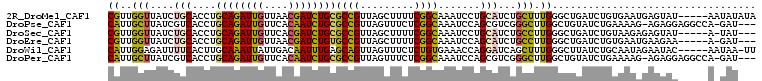
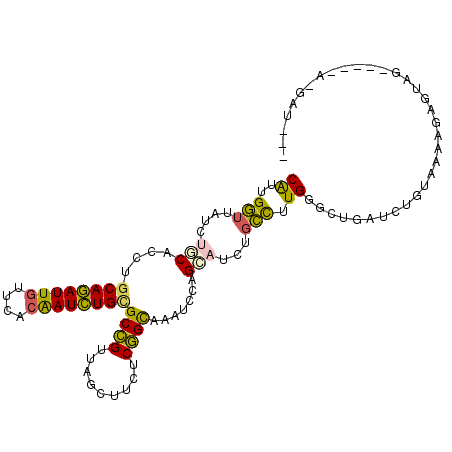
>2R_DroMel_CAF1 3081090 103 - 20766785 CGUUGGUUAUCUGCACCUGCAGAUUGUUAACGAUCUGCGCCGUUAGCUUUUCGGCAAAUCCUGCAUCUGCUUUGGGCUGAUCUGUGAAUGAGUAU-----AAUAUAUA .(((((..((((((....))))))..)))))((((.((((((.........))))....((.((....))...)))).)))).............-----........ ( -29.30) >DroPse_CAF1 391 103 - 1 CAUUGCUUAUCGUCACCUGCAGAUUGUUCACAAUCUGCGCCGUUAGUUUCUCGGCAAAUCCAGCGUCGGGCUUGGCUGUAUCUGAAAAG-AGAGGAGGCCA-GAU--- ....((((..(((.....((((((((....))))))))((((.........)))).......)))..))))((((((...(((....))-).....)))))-)..--- ( -35.80) >DroSec_CAF1 390 99 - 1 CGUUGGUUAUCUGCACCUGCAGAUUGUUCACGAUCUGCGCCGUUAGCUUUUCGGCAAAUCCUGCAUCUGCCUUGGGCUGAUCUGUAAGAGAGUAU-----A-UAU--- .........((((((...((((((((....))))))))...((((((((...((((...........))))..)))))))).))).)))......-----.-...--- ( -26.30) >DroEre_CAF1 395 99 - 1 CGUUGGUUAUCUGCACCUGCAGAUUGUUAACGAUCUGUGCCGUUAGCUUUUCGGCAAAUCCAGCAUCUGCCUUGGGCUGAUCUGUGAAUGAAGAA-----A-GAU--- ((((((..((((((....))))))..))))))((((.(((((.........)))))..(((((.(((.(((...))).)))))).))........-----)-)))--- ( -31.70) >DroWil_CAF1 383 102 - 1 CAUUGGAGAUUUUCACUUGCAAAUUAUUGACAAUUUGAGCAGUUAGUUUCUCUGUGAAACCAGGAUCAGCUUUGGGCUUAUCUGCAAUAGAAUAC-----AAUAA-UU .((((((((((...(((..((((((......))))))...))).))))))(((((.......((((.(((.....))).))))...)))))...)-----)))..-.. ( -18.30) >DroPer_CAF1 391 103 - 1 CAUUGCUUAUCGUCACCUGCAGAUUGUUCACAAUCUGCGCCGUUAGUUUCUCGGCAAAUCCAGCGUCGGGCUUGGCUGUAUCUGAAAAG-AGAGGAGGCCA-GAU--- ....((((..(((.....((((((((....))))))))((((.........)))).......)))..))))((((((...(((....))-).....)))))-)..--- ( -35.80) >consensus CAUUGGUUAUCUGCACCUGCAGAUUGUUCACAAUCUGCGCCGUUAGCUUCUCGGCAAAUCCAGCAUCUGCCUUGGGCUGAUCUGUAAAAGAGUAG_____A_GAU___ ((..(((....(((....((((((((....))))))))((((.........)))).......)))...))).)).................................. (-18.60 = -17.63 + -0.97)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:39 2006