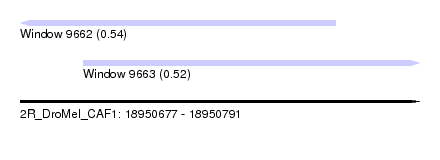
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,950,677 – 18,950,791 |
| Length | 114 |
| Max. P | 0.539095 |

| Location | 18,950,677 – 18,950,767 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.81 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -11.91 |
| Energy contribution | -12.05 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
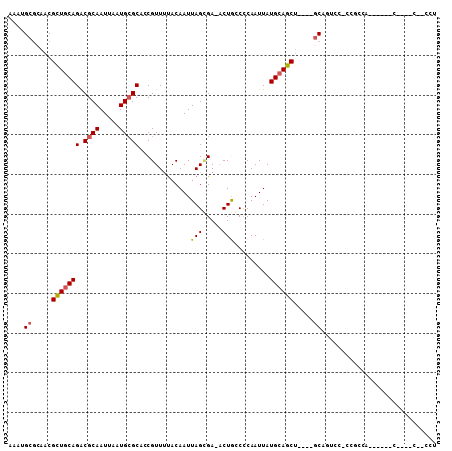
>2R_DroMel_CAF1 18950677 90 - 20766785 AAAUGCGCAACGCUGCAGACGCAAUUAAUGCGCACCGUUUUACAAUUAGCGA-CCUGGCCCAAUUAUGCAGCU----GCCACCC-CCUGCA------C----C--CCU ...((.(((..(((((((.((((.....)))))..((((........)))).-.............)))))))----))))...-......------.----.--... ( -19.40) >DroPse_CAF1 52560 93 - 1 AAAUGCGCAACGCUGCAGACGCAAUUAAUGCGCAACGUUUUACAAUUAGUGAAACUACCUCAAUUAUGCAGCU----GCAGUCC-CAGCCA------C----CAACCU ......(((..(((((((.((((.....)))))...(((((((.....)))))))...........)))))))----)).....-......------.----...... ( -23.30) >DroYak_CAF1 51580 83 - 1 AAAUGCGCAACGCUGCAGACGCAAUUAAUGCGCACCGUUUUACAAUUAGCGA-CCUGGCCCAAUUAUGCAGCU----GCA--CC-CCUGCC----------------- ......(((..(((((((.((((.....)))))..((((........)))).-.............)))))))----)).--..-......----------------- ( -19.50) >DroMoj_CAF1 72067 100 - 1 AAACGAGCUUCGUUACAGGCGCAAUUAAUGAGCUGCGUUCCACAAUUAGGGA-ACUGCCUCAAUUAUGCAGCCAACAACAGUCGCCGGCCACC----CCAGCC---CU .((((.....))))...(((((......((.(((((((........(((...-.)))........)))))))))......).))))(((....----...)))---.. ( -25.69) >DroAna_CAF1 58014 92 - 1 AAAUGCUAAACGCUGCAGACGCAAUUAAUGCGCACCGUUUUACAAUUAGCGA-ACUGGCCCAAUUAUGCAGCU----GCC--UU-CCUGCA--AAGCC----C--CCU ....(((....(((((((.((((.....)))))..((((........)))).-.............))))))(----((.--..-...)))--.))).----.--... ( -20.30) >DroPer_CAF1 53572 93 - 1 AAAUGCGCAACGCUGCAGACGCAAUUAAUGCGCAACGUUUUACAAUUAGUGAAACUACCUCAAUUAUGCAGCU----GCAGUCC-CAGCCA------C----CAACCU ......(((..(((((((.((((.....)))))...(((((((.....)))))))...........)))))))----)).....-......------.----...... ( -23.30) >consensus AAAUGCGCAACGCUGCAGACGCAAUUAAUGCGCACCGUUUUACAAUUAGCGA_ACUGCCCCAAUUAUGCAGCU____GCAGUCC_CCGCCA______C____C__CCU ....((.....(((((((.((((.....))))).............(((.....))).........)))))).....))............................. (-11.91 = -12.05 + 0.14)


| Location | 18,950,695 – 18,950,791 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -22.00 |
| Energy contribution | -21.50 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
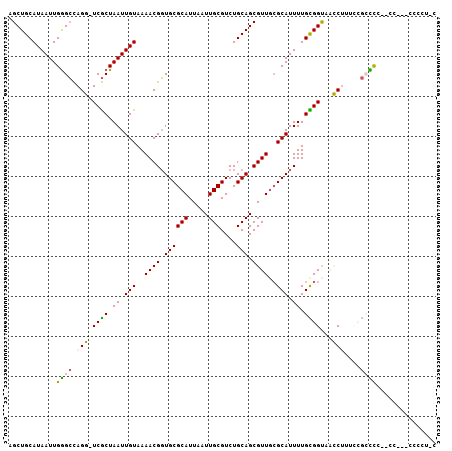
>2R_DroMel_CAF1 18950695 96 + 20766785 AGCUGCAUAAUUGGGCCAGG-UCGCUAAUUGUAAAACGGUGCGCAUUAAUUGCGUCUGCAGCGUUGCGCAUUUUGCGGCAACCUUUUCGCCUC--GCC--------C ............((((.(((-(.(((.((((.....))))(((((.....)))))....)))(((((((.....)).)))))......)))).--)))--------) ( -29.20) >DroPse_CAF1 52580 101 + 1 AGCUGCAUAAUUGAGGUAGUUUCACUAAUUGUAAAACGUUGCGCAUUAAUUGCGUCUGCAGCGUUGCGCAUUUUGUGGCUUGCC----ACUCGAGAG--UCGCCUCC ............((((((.((((......(((..(((((((((((.....)))....))))))))..)))....((((....))----))..)))).--).))))). ( -30.70) >DroSec_CAF1 49256 102 + 1 AGCUGCAUAAUUGGGCCAGG-UCGCUAAUUGUAAAACGGUGCGCAUUAAUUGCGUCUGCAGCGUUGCGCAUUUUGUGGUAACCUUUCCGCCCC--CC--UCCCCUUC ............((((.(((-(..(((..(((..((((.((((((.....)))....))).))))..))).....)))..))))....)))).--..--........ ( -28.00) >DroSim_CAF1 52577 104 + 1 AGCUGCAUAAUUGGGCCAGG-UCGCUAAUUGUAAAACGGUGCGCAUUAAUUGCGUCUGCAGCGUUGCGCAUUUUGCGGUAACCUUUCCGCCCC--CCCCACCCUUUC ............((((.(((-(..((....(((((...(((((((....((((....))))...))))))))))))))..))))....)))).--............ ( -28.00) >DroYak_CAF1 51591 104 + 1 AGCUGCAUAAUUGGGCCAGG-UCGCUAAUUGUAAAACGGUGCGCAUUAAUUGCGUCUGCAGCGUUGCGCAUUUUGCGGUUACCCUUCCGACCU--UCCCACCGCACC ...(((.....((((..(((-(((.((((((((((...(((((((....((((....))))...)))))))))))))))))......))))))--.))))..))).. ( -33.50) >DroAna_CAF1 58034 87 + 1 AGCUGCAUAAUUGGGCCAGU-UCGCUAAUUGUAAAACGGUGCGCAUUAAUUGCGUCUGCAGCGUUUAGCAUUUUGUGGUUUGCU----GGUC--------------- .............(((((((-..((((..(((.(((((.((((((.....)))....))).))))).))).....))))..)))----))))--------------- ( -27.60) >consensus AGCUGCAUAAUUGGGCCAGG_UCGCUAAUUGUAAAACGGUGCGCAUUAAUUGCGUCUGCAGCGUUGCGCAUUUUGCGGUAACCUUUCCGCCCC__CC___CCCCU_C ............((((.(((.((((.((.(((..((((.((((((.....)))....))).))))..))).)).))))...)))....))))............... (-22.00 = -21.50 + -0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:44 2006