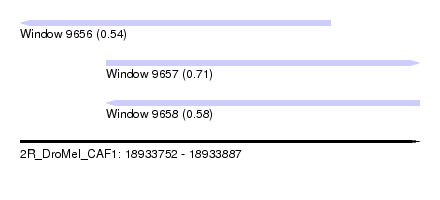
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,933,752 – 18,933,887 |
| Length | 135 |
| Max. P | 0.705746 |

| Location | 18,933,752 – 18,933,857 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 87.52 |
| Mean single sequence MFE | -27.04 |
| Consensus MFE | -20.22 |
| Energy contribution | -20.02 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18933752 105 - 20766785 AGCUAACAU----UCACAUUGCAAUUUUACUGAUAUUAUAACAGCCCGCGAGAUCUCUUACGCUGUUACAUGAACCGCGGGAAAGCAGAGAGUGAAAGAAGUGUAUUAU .(((....(----((((.((((.......(((.........)))((((((.(.((................)).)))))))...))))...)))))...)))....... ( -20.29) >DroSec_CAF1 32865 105 - 1 AGCUAACAU----UCACAUUGCAAUUUUGCUGAUAUUGUAACAGCACGCGAGAUCUCUUACGCUGUUACAUGAACCGCGGGAAAGCAGAGAGUGAAAGAAGUGUACUAU .(((....(----((((.((((..((((((.(....(((((((((..(..((....))..))))))))))....).))))))..))))...)))))...)))....... ( -26.70) >DroSim_CAF1 35860 105 - 1 AGCUAACAU----UCACAUUGCAAUUUUGCUGAUAUUGUAACAGCACGCGAGAUCUCUUACGCUGUUACAUGAACCGCGGGAAAGCAGAGAGUGAAAGAAGUGUACUAU .(((....(----((((.((((..((((((.(....(((((((((..(..((....))..))))))))))....).))))))..))))...)))))...)))....... ( -26.70) >DroEre_CAF1 33779 104 - 1 AGCUAACGU----ACACAUUGCAAUUUUGCU-AUAUUGUAACAGAACGAGCGAUCUCUUCCUCUGUUACAUGAACAGCGGAGAGGCUGAGAGUGAAAGAAGUGCACUUU .((....))----......((((.(((..((-....(((((((((..(((......)))..)))))))))....((((......))))..))..)))....)))).... ( -26.80) >DroYak_CAF1 34521 109 - 1 AUCCAACGUACGUACCCAUUGCAAUUUUGCUGAUAUUGUAACAGCUAGCGAGAUCUCUCACUCUGUUACAUUAACAGCGAGAAAGCAGAGAGUGAAAGAAGUGCACUAU .......((((.(((.(.((((..((((((((.((.((((((((..((.(((....))).)))))))))).)).))))))))..)))).).)))......))))..... ( -34.70) >consensus AGCUAACAU____UCACAUUGCAAUUUUGCUGAUAUUGUAACAGCACGCGAGAUCUCUUACGCUGUUACAUGAACCGCGGGAAAGCAGAGAGUGAAAGAAGUGUACUAU ...................((((.(((((((.....(((((((((..(.(((....))).))))))))))......((......))....)))))))....)))).... (-20.22 = -20.02 + -0.20)

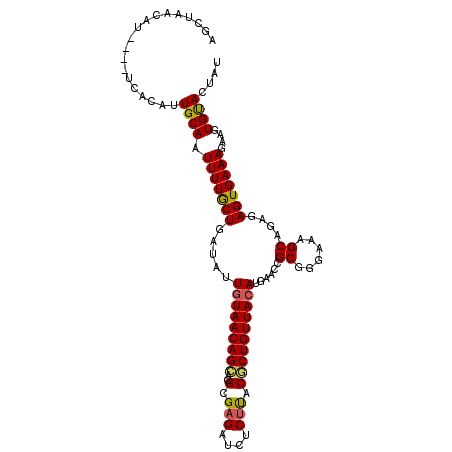
| Location | 18,933,781 – 18,933,887 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.32 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.02 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18933781 106 + 20766785 CCGCGGUUCAUGUAACAGCGUAAGAGAUCUCGCGGGCUGUUAUAAUAUCAGUAAAAUUGCAAUGUGA----AUGUUAGCUGAUGCAAAAGAGAAAGGAGAGAGCAGGAAC---------- ((...((((.(((((((((.(..((....))..).))))))))).(((((((.((....(.....).----...)).)))))))................)))).))...---------- ( -25.80) >DroSec_CAF1 32894 106 + 1 CCGCGGUUCAUGUAACAGCGUAAGAGAUCUCGCGUGCUGUUACAAUAUCAGCAAAAUUGCAAUGUGA----AUGUUAGCUGAUGCAAAAGAGAAAGGAGAGAGCAGGAAG---------- ((...((((.((((((((((...((....))...)))))))))).(((((((.((....(.....).----...)).)))))))................)))).))...---------- ( -29.00) >DroSim_CAF1 35889 106 + 1 CCGCGGUUCAUGUAACAGCGUAAGAGAUCUCGCGUGCUGUUACAAUAUCAGCAAAAUUGCAAUGUGA----AUGUUAGCUGAUGCAAAAGAGAAUGGAGAGAGCAGGAAC---------- ((...((((.((((((((((...((....))...)))))))))).(((((((.((....(.....).----...)).)))))))................)))).))...---------- ( -29.00) >DroEre_CAF1 33808 113 + 1 CCGCUGUUCAUGUAACAGAGGAAGAGAUCGCUCGUUCUGUUACAAUAU-AGCAAAAUUGCAAUGUGU----ACGUUAGCUGAUGCAAAAGAGAAAGGAGGGAGCAAUAAC--UACUUUCG ..(((((...((((((((((...(((....))).))))))))))..))-)))....(((((..((..----......))...)))))....(((((.((..(....)..)--).))))). ( -32.30) >DroYak_CAF1 34550 112 + 1 UCGCUGUUAAUGUAACAGAGUGAGAGAUCUCGCUAGCUGUUACAAUAUCAGCAAAAUUGCAAUGGGUACGUACGUUGGAUGUUGCAGAAGAGAAAGGAGAGAGCAAUAACUA-------- ..((((.((.((((((((((((((....))))))..)))))))).)).)))).......(((((........)))))..((((((.................))))))....-------- ( -32.73) >consensus CCGCGGUUCAUGUAACAGCGUAAGAGAUCUCGCGUGCUGUUACAAUAUCAGCAAAAUUGCAAUGUGA____AUGUUAGCUGAUGCAAAAGAGAAAGGAGAGAGCAGGAAC__________ .....((((.(((((((((((.((....)).))..)))))))))......(((.....(((((((......))))).))...)))...............))))................ (-18.14 = -18.02 + -0.12)

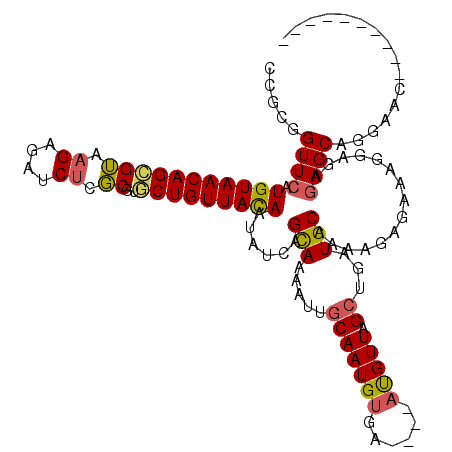
| Location | 18,933,781 – 18,933,887 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.32 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -15.98 |
| Energy contribution | -15.98 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
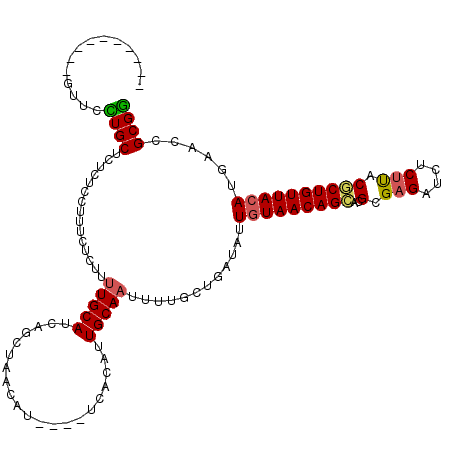
>2R_DroMel_CAF1 18933781 106 - 20766785 ----------GUUCCUGCUCUCUCCUUUCUCUUUUGCAUCAGCUAACAU----UCACAUUGCAAUUUUACUGAUAUUAUAACAGCCCGCGAGAUCUCUUACGCUGUUACAUGAACCGCGG ----------....((((...................(((((.(((.((----((.....).))).))))))))....(((((((..(..((....))..))))))))........)))) ( -17.00) >DroSec_CAF1 32894 106 - 1 ----------CUUCCUGCUCUCUCCUUUCUCUUUUGCAUCAGCUAACAU----UCACAUUGCAAUUUUGCUGAUAUUGUAACAGCACGCGAGAUCUCUUACGCUGUUACAUGAACCGCGG ----------....((((...................((((((......----...............))))))..(((((((((..(..((....))..))))))))))......)))) ( -23.70) >DroSim_CAF1 35889 106 - 1 ----------GUUCCUGCUCUCUCCAUUCUCUUUUGCAUCAGCUAACAU----UCACAUUGCAAUUUUGCUGAUAUUGUAACAGCACGCGAGAUCUCUUACGCUGUUACAUGAACCGCGG ----------....((((...................((((((......----...............))))))..(((((((((..(..((....))..))))))))))......)))) ( -23.70) >DroEre_CAF1 33808 113 - 1 CGAAAGUA--GUUAUUGCUCCCUCCUUUCUCUUUUGCAUCAGCUAACGU----ACACAUUGCAAUUUUGCU-AUAUUGUAACAGAACGAGCGAUCUCUUCCUCUGUUACAUGAACAGCGG .(((((.(--(..........)).)))))....(((((...((....))----......)))))...((((-....(((((((((..(((......)))..))))))))).....)))). ( -22.80) >DroYak_CAF1 34550 112 - 1 --------UAGUUAUUGCUCUCUCCUUUCUCUUCUGCAACAUCCAACGUACGUACCCAUUGCAAUUUUGCUGAUAUUGUAACAGCUAGCGAGAUCUCUCACUCUGUUACAUUAACAGCGA --------..........................(((((......((....)).....)))))...((((((.((.((((((((..((.(((....))).)))))))))).)).)))))) ( -25.10) >consensus __________GUUCCUGCUCUCUCCUUUCUCUUUUGCAUCAGCUAACAU____UCACAUUGCAAUUUUGCUGAUAUUGUAACAGCACGCGAGAUCUCUUACGCUGUUACAUGAACCGCGG ..............((((...............(((((.....................)))))............(((((((((..(.(((....))).))))))))))......)))) (-15.98 = -15.98 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:39 2006