
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,929,798 – 18,929,897 |
| Length | 99 |
| Max. P | 0.772543 |

| Location | 18,929,798 – 18,929,897 |
|---|---|
| Length | 99 |
| Sequences | 6 |
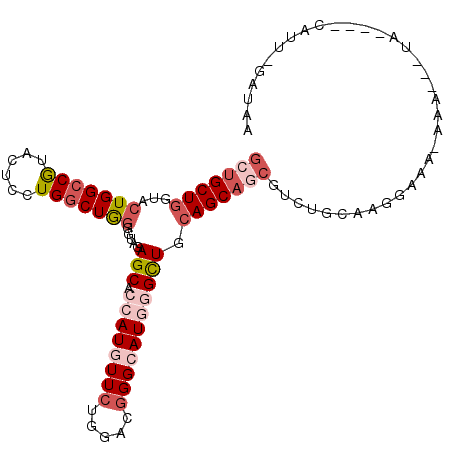
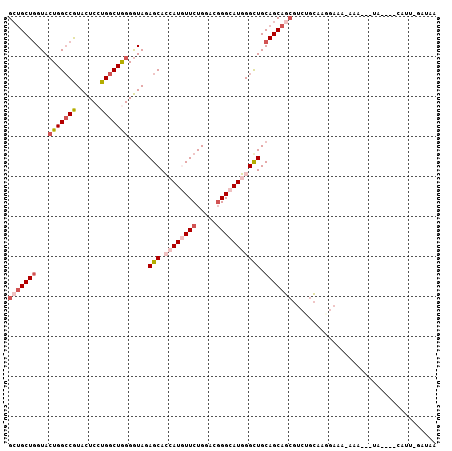
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 79.69 |
| Mean single sequence MFE | -35.55 |
| Consensus MFE | -25.01 |
| Energy contribution | -26.93 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18929798 99 + 20766785 GCUGCUGGUACUGGCCGUACUCCUGGCUGGGGUACAGCACCAUGUUCUGCACGGGCAUGGGCUGCAGCAGCGUCUGCAAUCAAAGAAA---UA----CGUU-GAUAA (((((((...(..((((......))))..)....((((.((((((((.....))))))))))))))))))).......(((((.(...---..----).))-))).. ( -41.10) >DroVir_CAF1 43857 103 + 1 GACGCUCGUAGUGGGCAUAUUCCUGACUAGGAUAGAGCAGCAUAUUCUGUACGGGCAUCGGCUGCAGCAGAGUCUGCAAGG-AAUCAACAGUA--GAUAUU-UAAAA ...(((.(((((.((......((((((.((((((........)))))))).))))..)).))))))))...((((((....-........)))--)))...-..... ( -24.80) >DroPse_CAF1 29068 103 + 1 GCUGCUGGUACUGGCCGUACUCGUGGCUGGGGUAGAGCAGCAUAUUCUGCACUGGCAUCGGUUGCAGCAUCGUCUGAAAAGAACGGAA---UUCAGAUUUU-AAUUU ((((((.(..(..((((......))))..)..)..)))))).....((((((((....))).)))))....(((((((..........---)))))))...-..... ( -33.00) >DroSec_CAF1 28965 100 + 1 GCUGCUGGUACUGGCCGUACUCCUGGCUGGGGUAGAGCACCAUGUUCUGGACGGGCAUGGGCUGCAGCAGCGUCUGCAAGGAAG-AAAAG-AA----CAUU-GAUAA (((((((...(..((((......))))..).....(((.((((((((.....))))))))))).))))))).(((.......))-)....-..----....-..... ( -37.80) >DroSim_CAF1 31976 98 + 1 GCUGCUGGUACUGGCCGUACUCCUGGCUGGGGUAGAGCACCAUGUUCUGGACGGGCAUGGGCUGCAGCAGCGUCUGCAAGGAAA-AAA---UA----UAUU-GGUAC (((((((...(..((((......))))..).....(((.((((((((.....))))))))))).))))))).(((....)))..-...---..----....-..... ( -37.50) >DroEre_CAF1 29916 102 + 1 GCUGCUGGUACUGGCCGUACUCCUGGCUGGGGUAGAGCACCAUGUUCUGGACGGGCAUGGGCUGCAGCAGCGUCUGCAACGAUACCAAAG-GC----CAUUUAAUUU (((((((...(..((((......))))..).....(((.((((((((.....))))))))))).)))))))((((.............))-))----.......... ( -39.12) >consensus GCUGCUGGUACUGGCCGUACUCCUGGCUGGGGUAGAGCACCAUGUUCUGGACGGGCAUGGGCUGCAGCAGCGUCUGCAAGGAAA_AAA___UA____CAUU_GAUAA (((((((...(((((((......))))))).....(((.((((((((.....))))))))))).))))))).................................... (-25.01 = -26.93 + 1.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:35 2006