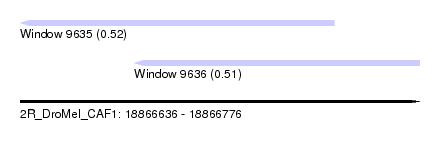
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,866,636 – 18,866,776 |
| Length | 140 |
| Max. P | 0.521576 |

| Location | 18,866,636 – 18,866,746 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.32 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -19.43 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
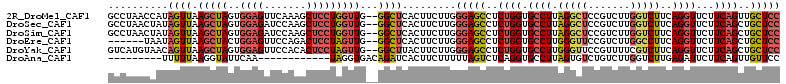
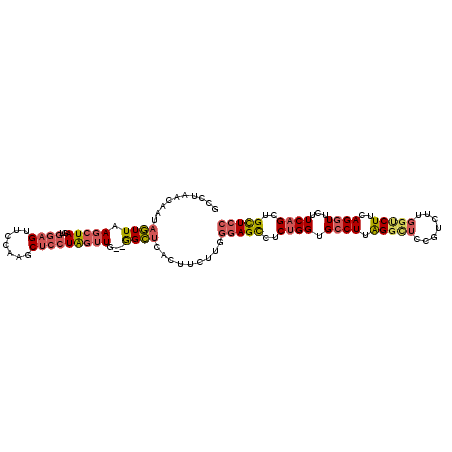
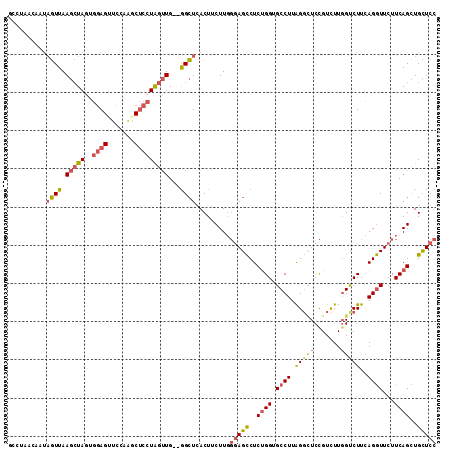
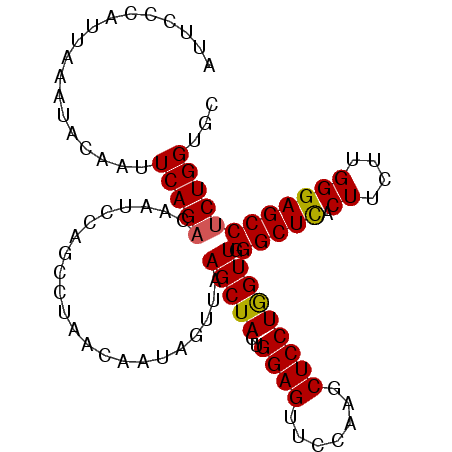
>2R_DroMel_CAF1 18866636 110 - 20766785 GCCUAACCAUAGUUAAGCUAGUGGAGUUCAAAGCUCCUGGUUG--GGCUCACUUCUUGGGAGCCUCUGGUGCCUUAGGCUCCGUCUUGGUCUUCAGGUUCUUCAGUUGCUCC ((((((((((((.....)))..(((((.....)))))))))))--)))..........(((((..((((.((((.(((((.......)))))..))))...))))..))))) ( -43.20) >DroSec_CAF1 209 110 - 1 GCCUAACUAUAGUUAAGCUAGUGGAGAUCCAAGCUCCUGGUUG--GGCUCACUUCUUGGGAGCCUCUGGUGCCUUAGGCUCCGUCUUGGUCUUCAGGUUCUUCAGCUGCUCC ((((((((((((.....)))..((((.......))))))))))--)))..........(((((..((((.((((.(((((.......)))))..))))...))))..))))) ( -37.80) >DroSim_CAF1 209 110 - 1 GCCUAACUAUAGUUAAGCUAGUGGAGAUCCAAGCUCCUGGUUG--GGCUCACUUCUUGGGAGCCUCUGGUGCCUUAGGCUCCGUCUUGGUCUUCAGGUUCUUCAGCUGCUCC ((((((((((((.....)))..((((.......))))))))))--)))..........(((((..((((.((((.(((((.......)))))..))))...))))..))))) ( -37.80) >DroEre_CAF1 671 104 - 1 ------UAAUAGUUAAGCUACUGGAGUUCCAGACUCCUAGUUG--GGCUCACUUCUUGGGAGCCUCUGGUGCCUUGGGUUCCGUCUUGGCCUUCAGGUUCUUCAGCUGCUCC ------.........(((..((((((..((((((((((((..(--(.....))..)))))))..))))).((((.(((((.......)))))..)))).))))))..))).. ( -33.40) >DroYak_CAF1 213 110 - 1 GUCAUGUAACAGUUAAGCUAGUGGAGUUCCACACUCCUAGUUG--GGCUUACUUCUUGGGAGCCUCUGGUGCCUUGGGUUCCGUUUUCGUCUUCAGGUUCUUCAGCUGCUCC .....(((..(((..(((((..(((((.....)))))))))).--.))))))......(((((..((((.((((..((...((....)).))..))))...))))..))))) ( -30.80) >DroAna_CAF1 11335 91 - 1 ---------UUUUUAAGGUAUUCAA------------UAGGUGACAGAUCACUUCUUUUUAGUCUCAGGUGCCUUAGUGUCUGUCUUGGUCUUGAGAUUCUUCAGUUGUUCC ---------.......((....(((------------(((((((....))))))......(((((((((.(((..((.......)).)))))))))))).....))))..)) ( -18.60) >consensus GCCUAACAAUAGUUAAGCUAGUGGAGUUCCAAGCUCCUAGUUG__GGCUCACUUCUUGGGAGCCUCUGGUGCCUUAGGCUCCGUCUUGGUCUUCAGGUUCUUCAGCUGCUCC ..........((((.(((((..((((.......)))))))))...)))).........(((((..((((.((((.(((((.......)))))..))))...))))..))))) (-19.43 = -20.20 + 0.77)
| Location | 18,866,676 – 18,866,776 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 86.10 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.06 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18866676 100 - 20766785 AUUCCCAUUAAAUACAAUUCAGACAAUCCAGCCUAACCAUAGUUAAGCUAGUGGAGUUCAAAGCUCCUGGUUGGGCUCACUUCUUGGGAGCCUCUGGUGC ..................(((((...((((((((((((((((.....)))..(((((.....)))))))))))))))..(.....))))...)))))... ( -30.10) >DroSec_CAF1 249 100 - 1 AUUCAUAUUAAAUACAAUUCAGACAAUCCAGCCUAACUAUAGUUAAGCUAGUGGAGAUCCAAGCUCCUGGUUGGGCUCACUUCUUGGGAGCCUCUGGUGC ..................((((((((((.(((.((((....)))).)))...((((.......)))).)))))(((((.((....))))))))))))... ( -26.80) >DroSim_CAF1 249 100 - 1 AUUCGUAUUAAAUACAAUUCAGACAAUCCAGCCUAACUAUAGUUAAGCUAGUGGAGAUCCAAGCUCCUGGUUGGGCUCACUUCUUGGGAGCCUCUGGUGC ....((((...))))...((((((((((.(((.((((....)))).)))...((((.......)))).)))))(((((.((....))))))))))))... ( -27.20) >DroEre_CAF1 711 86 - 1 AUUCCCAUUAAAUACAAUUCAG--------------UAAUAGUUAAGCUACUGGAGUUCCAGACUCCUAGUUGGGCUCACUUCUUGGGAGCCUCUGGUGC .................(((((--------------((..........)))))))((.((((((((((((..((.....))..)))))))..))))).)) ( -22.10) >DroYak_CAF1 253 100 - 1 AUUCCCAUUAAAUACAAUUCAGACAAUGCAGUCAUGUAACAGUUAAGCUAGUGGAGUUCCACACUCCUAGUUGGGCUUACUUCUUGGGAGCCUCUGGUGC ..(((((..............(((......)))..(((..(((..(((((..(((((.....))))))))))..))))))....)))))(((...))).. ( -23.00) >consensus AUUCCCAUUAAAUACAAUUCAGACAAUCCAGCCUAACAAUAGUUAAGCUAGUGGAGUUCCAAGCUCCUGGUUGGGCUCACUUCUUGGGAGCCUCUGGUGC ..................(((((......................(((((..((((.......))))))))).(((((.((....))))))))))))... (-19.26 = -19.06 + -0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:17 2006