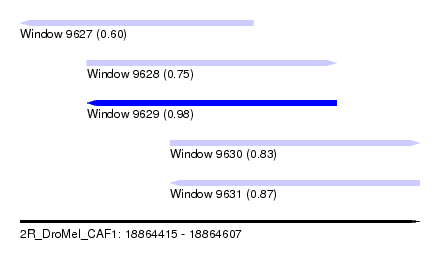
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,864,415 – 18,864,607 |
| Length | 192 |
| Max. P | 0.979117 |

| Location | 18,864,415 – 18,864,527 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.38 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -21.51 |
| Energy contribution | -22.66 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
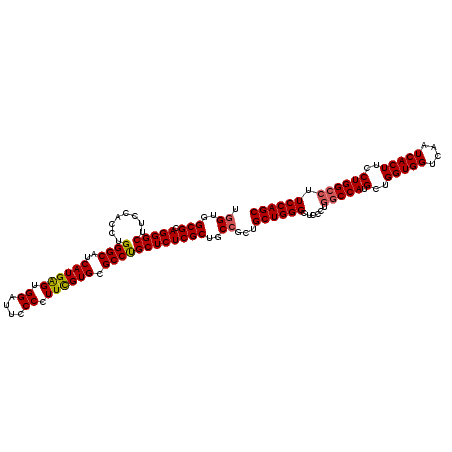
>2R_DroMel_CAF1 18864415 112 - 20766785 CUGGCCAUGCUGGUGGUCAAUCACUUCCUGGCCUUCCAGCAUUUCGUCACUGUCUAUGUGCCUUUCACGCAGGUGAGCAU--------AUGUAUUGCAUAAAUCAUUAGUCAUGGAAAGC .((((.(((((((.(((((.........)))))..)))))))...))))((.(((((((((...((((....)))))))(--------(((.....))))..........)))))).)). ( -31.80) >DroSec_CAF1 39823 112 - 1 CUGGCCAUGCUGGUGGUCAAUCACUUCCUGGCCUUCCAGCACUUCACCACCGUCUAUGUGCCUUUCACGCAGGUGAGCAU--------AUGUAUUGCAUAAAUCAUGAAUCAUGGAAAGC ((..(((((((((.(((((.........)))))..)))))..((((.((((.....((((.....))))..)))).(((.--------......)))........))))..))))..)). ( -30.30) >DroSim_CAF1 39240 112 - 1 CUGGCCAUGCUGGUGGUCAAUCACUUCCUGGCCUUCCAGCACUUCACAACCGUCUAUGUGCCUUUCACGCAGGUGAGCAU--------AUGUAUUGCAUAAAUCAUGAAUCAUGGAAAGC ((..(((((((((.(((((.........)))))..)))))..((((.....(..(((.(((.......))).)))..).(--------(((.....)))).....))))..))))..)). ( -29.60) >DroEre_CAF1 42047 104 - 1 UUGGCCAUGCUGGUGGCCAAUCACUUCCUGGCCUUCCAGCACUUCACUUCCGUCUACGUGCCUUUCACGCAGGUGAGCCUAUUACCUUAUUCGCUACGUAUAUA---------------- ..(((..((((((.(((((.........)))))..))))))..((((((.(((.............))).))))))))).........................---------------- ( -29.32) >DroYak_CAF1 41302 120 - 1 UUGACCAUGCUGGUGGUCAAUCACUUCCUGGCAUUCCAGCACUUCACCACUGUCUAUGUGCCUUUCACCCAGGUGAGCCCAUCCCCUUAUGUAUUGCAUAAAUCAUUAGUCAUGGAAAGC ....(((((((((((((............(((......((((...............))))...((((....))))))).......(((((.....))))))))))))).)))))..... ( -30.56) >consensus CUGGCCAUGCUGGUGGUCAAUCACUUCCUGGCCUUCCAGCACUUCACCACCGUCUAUGUGCCUUUCACGCAGGUGAGCAU________AUGUAUUGCAUAAAUCAUGAAUCAUGGAAAGC ....(((((((((.(((((.........)))))..)))))..............(((((.....((((....))))(((((......)))))...)))))...........))))..... (-21.51 = -22.66 + 1.15)

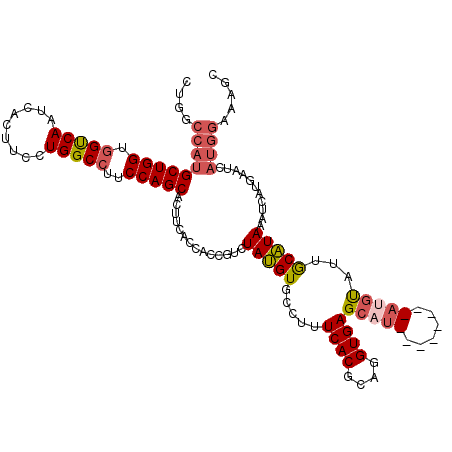
| Location | 18,864,447 – 18,864,567 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -45.50 |
| Consensus MFE | -34.60 |
| Energy contribution | -35.76 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18864447 120 + 20766785 AUGCUCACCUGCGUGAAAGGCACAUAGACAGUGACGAAAUGCUGGAAGGCCAGGAAGUGAUUGACCACCAGCAUGGCCAGGGAGCCCAGCAGCGGCAGCGAGAGCAGGCGCACGAAGGGG .(((((.((((((((.....)))...........((...((((((..(((((.(..(((......)))...).))))).(....))))))).)))))).).)))))......(....).. ( -39.90) >DroSec_CAF1 39855 120 + 1 AUGCUCACCUGCGUGAAAGGCACAUAGACGGUGGUGAAGUGCUGGAAGGCCAGGAAGUGAUUGACCACCAGCAUGGCCAGGGAGCCCAGCAGAGGCAGCGAGAGCGGGCGCACGAAGGGA .......(((.((((...(((.(((.(..((((((.((.(.((((....)))).).....)).))))))..))))))).....((((.((....)).((....)))))).)))).))).. ( -48.30) >DroSim_CAF1 39272 120 + 1 AUGCUCACCUGCGUGAAAGGCACAUAGACGGUUGUGAAGUGCUGGAAGGCCAGGAAGUGAUUGACCACCAGCAUGGCCAGGGAGCCCAGCAGAGGCAGCGAGAGCAGGCGCACGAAGGGA .......(((.((((...(((.(((((....)))))..(((((((..((.(((.......))).)).))))))).))).....(((..((....)).((....)).))).)))).))).. ( -39.80) >DroEre_CAF1 42071 120 + 1 AGGCUCACCUGCGUGAAAGGCACGUAGACGGAAGUGAAGUGCUGGAAGGCCAGGAAGUGAUUGGCCACCAGCAUGGCCAAGGAGCCCAGCAGCGGCAGCGAGAGCAGGCGCACGAAGGGG .(((((..(((((((.....)))))))((....))(..(((((((..((((((.......)))))).)))))))..)....))))).....(((.(.((....)).).))).(....).. ( -52.60) >DroYak_CAF1 41342 120 + 1 GGGCUCACCUGGGUGAAAGGCACAUAGACAGUGGUGAAGUGCUGGAAUGCCAGGAAGUGAUUGACCACCAGCAUGGUCAAGGAGCCCAGCAGCGGCAGCGAGAGCAGGCGCACAAAGGGG ((((((.((((((((.....))).....((((........)))).....)))))......(((((((......))))))).))))))....(((.(.((....)).).)))......... ( -46.90) >consensus AUGCUCACCUGCGUGAAAGGCACAUAGACGGUGGUGAAGUGCUGGAAGGCCAGGAAGUGAUUGACCACCAGCAUGGCCAGGGAGCCCAGCAGCGGCAGCGAGAGCAGGCGCACGAAGGGG .......(((.((((...(((.(((.(..((((((.((.(.((((....)))).).....)).))))))..))))))).....(((..((....)).((....)).))).)))).))).. (-34.60 = -35.76 + 1.16)

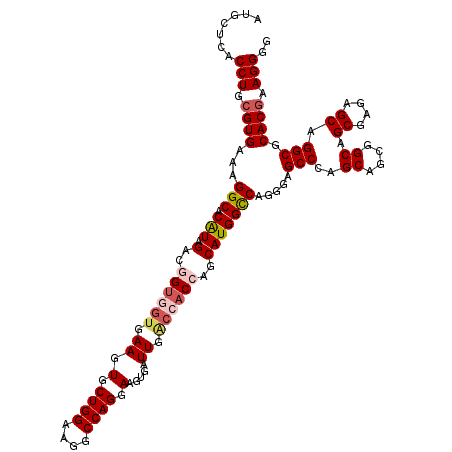
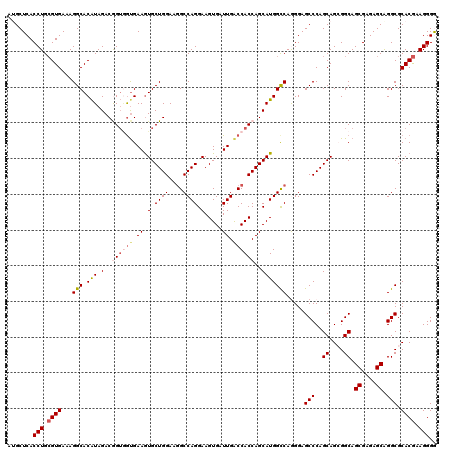
| Location | 18,864,447 – 18,864,567 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -39.02 |
| Consensus MFE | -34.92 |
| Energy contribution | -35.00 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18864447 120 - 20766785 CCCCUUCGUGCGCCUGCUCUCGCUGCCGCUGCUGGGCUCCCUGGCCAUGCUGGUGGUCAAUCACUUCCUGGCCUUCCAGCAUUUCGUCACUGUCUAUGUGCCUUUCACGCAGGUGAGCAU .......((.(((((((....((....)).(((((((....((((.(((((((.(((((.........)))))..)))))))...))))..))))).)).........))))))).)).. ( -40.20) >DroSec_CAF1 39855 120 - 1 UCCCUUCGUGCGCCCGCUCUCGCUGCCUCUGCUGGGCUCCCUGGCCAUGCUGGUGGUCAAUCACUUCCUGGCCUUCCAGCACUUCACCACCGUCUAUGUGCCUUUCACGCAGGUGAGCAU (((((.((((.((((((....)).((....)).)))).....(((((((.(((((((..........((((....))))......)))))))..)))).)))...)))).))).)).... ( -37.59) >DroSim_CAF1 39272 120 - 1 UCCCUUCGUGCGCCUGCUCUCGCUGCCUCUGCUGGGCUCCCUGGCCAUGCUGGUGGUCAAUCACUUCCUGGCCUUCCAGCACUUCACAACCGUCUAUGUGCCUUUCACGCAGGUGAGCAU .......((.(((((((.......((((.....)))).....(((((((((((.(((((.........)))))..))))).....((....))..))).)))......))))))).)).. ( -38.80) >DroEre_CAF1 42071 120 - 1 CCCCUUCGUGCGCCUGCUCUCGCUGCCGCUGCUGGGCUCCUUGGCCAUGCUGGUGGCCAAUCACUUCCUGGCCUUCCAGCACUUCACUUCCGUCUACGUGCCUUUCACGCAGGUGAGCCU .......(((.((..((....)).)))))....((((((((((....((((((.(((((.........)))))..))))))...............((((.....)))))))).)))))) ( -41.20) >DroYak_CAF1 41342 120 - 1 CCCCUUUGUGCGCCUGCUCUCGCUGCCGCUGCUGGGCUCCUUGACCAUGCUGGUGGUCAAUCACUUCCUGGCAUUCCAGCACUUCACCACUGUCUAUGUGCCUUUCACCCAGGUGAGCCC .......(((.((..((....)).)))))....((((((.((((((((....))))))))......(((((.......((((...............)))).......))))).)))))) ( -37.30) >consensus CCCCUUCGUGCGCCUGCUCUCGCUGCCGCUGCUGGGCUCCCUGGCCAUGCUGGUGGUCAAUCACUUCCUGGCCUUCCAGCACUUCACCACCGUCUAUGUGCCUUUCACGCAGGUGAGCAU .......((.(((((((....((((.....((..((.....(((((((....))))))).......))..))....)))).................(((.....)))))))))).)).. (-34.92 = -35.00 + 0.08)

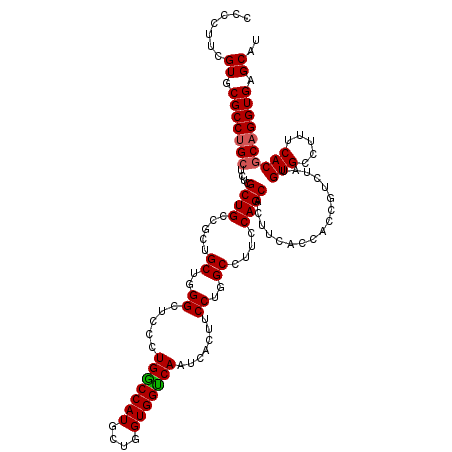

| Location | 18,864,487 – 18,864,607 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -50.00 |
| Consensus MFE | -44.05 |
| Energy contribution | -44.17 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18864487 120 + 20766785 GCUGGAAGGCCAGGAAGUGAUUGACCACCAGCAUGGCCAGGGAGCCCAGCAGCGGCAGCGAGAGCAGGCGCACGAAGGGGAAUCCACUCAUGAUGCCCAGGUGGAAGCCCUGCGCCACCA ((((...(((((.(..(((......)))...).))))).((....))..))))((..((....)).((((((.....(((..((((((..((.....))))))))..))))))))).)). ( -50.50) >DroSec_CAF1 39895 120 + 1 GCUGGAAGGCCAGGAAGUGAUUGACCACCAGCAUGGCCAGGGAGCCCAGCAGAGGCAGCGAGAGCGGGCGCACGAAGGGAAAUCCACUCAUGAUGCCCAGGUGGAAGCCCUGCGCCACCA (((((..((.(((.......))).)).))))).(((((((((...(((.(.......((....))(((((((.((..((....))..)).)).))))).).)))...))))).))))... ( -51.40) >DroSim_CAF1 39312 120 + 1 GCUGGAAGGCCAGGAAGUGAUUGACCACCAGCAUGGCCAGGGAGCCCAGCAGAGGCAGCGAGAGCAGGCGCACGAAGGGAAAUCCACUCAUGAUGCCCAGGUGGAAGCCCUGCGCCACCA (((((..(((((.(..(((......)))...).))))).(....))))))...((..((....)).((((((....(((...((((((..((.....))))))))..))))))))).)). ( -48.60) >DroYak_CAF1 41382 120 + 1 GCUGGAAUGCCAGGAAGUGAUUGACCACCAGCAUGGUCAAGGAGCCCAGCAGCGGCAGCGAGAGCAGGCGCACAAAGGGGAAUCCACCCAUGAUGCCCAGGUGGAAGCCCUGCGCCACGA ((((...(((..((......(((((((......)))))))....))..))).)))).((....)).((((((.....(((..((((((..((.....))))))))..))))))))).... ( -49.50) >consensus GCUGGAAGGCCAGGAAGUGAUUGACCACCAGCAUGGCCAGGGAGCCCAGCAGAGGCAGCGAGAGCAGGCGCACGAAGGGAAAUCCACUCAUGAUGCCCAGGUGGAAGCCCUGCGCCACCA (((((..(((((.(..(((......)))...).))))).(....)))))).......((....)).((((((....(((...((((((..((.....))))))))..))))))))).... (-44.05 = -44.17 + 0.13)

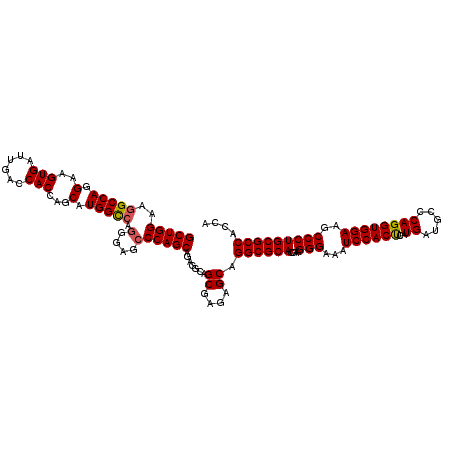
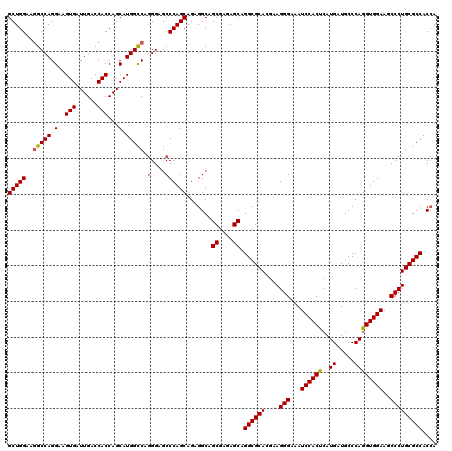
| Location | 18,864,487 – 18,864,607 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -50.75 |
| Consensus MFE | -46.09 |
| Energy contribution | -46.27 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18864487 120 - 20766785 UGGUGGCGCAGGGCUUCCACCUGGGCAUCAUGAGUGGAUUCCCCUUCGUGCGCCUGCUCUCGCUGCCGCUGCUGGGCUCCCUGGCCAUGCUGGUGGUCAAUCACUUCCUGGCCUUCCAGC .((..(((.(((((........((((..((((((.((....)).)))))).))))))))))))..))...((((((......(((((.(..(((((....)))))..)))))).)))))) ( -50.00) >DroSec_CAF1 39895 120 - 1 UGGUGGCGCAGGGCUUCCACCUGGGCAUCAUGAGUGGAUUUCCCUUCGUGCGCCCGCUCUCGCUGCCUCUGCUGGGCUCCCUGGCCAUGCUGGUGGUCAAUCACUUCCUGGCCUUCCAGC ..(((((.(((((.........((((..((((((.((....)).)))))).))))((((..((.......)).)))).))))))))))(((((.(((((.........)))))..))))) ( -52.60) >DroSim_CAF1 39312 120 - 1 UGGUGGCGCAGGGCUUCCACCUGGGCAUCAUGAGUGGAUUUCCCUUCGUGCGCCUGCUCUCGCUGCCUCUGCUGGGCUCCCUGGCCAUGCUGGUGGUCAAUCACUUCCUGGCCUUCCAGC ..(((((.(((((..((((.(.(((((.((((((.((....)).)))))).....((....)))))))..).))))..))))))))))(((((.(((((.........)))))..))))) ( -50.40) >DroYak_CAF1 41382 120 - 1 UCGUGGCGCAGGGCUUCCACCUGGGCAUCAUGGGUGGAUUCCCCUUUGUGCGCCUGCUCUCGCUGCCGCUGCUGGGCUCCUUGACCAUGCUGGUGGUCAAUCACUUCCUGGCAUUCCAGC ..(((((((((((..((((((((.......))))))))..))).....)))))).))....((((....(((..((....((((((((....))))))))......))..)))...)))) ( -50.00) >consensus UGGUGGCGCAGGGCUUCCACCUGGGCAUCAUGAGUGGAUUCCCCUUCGUGCGCCUGCUCUCGCUGCCGCUGCUGGGCUCCCUGGCCAUGCUGGUGGUCAAUCACUUCCUGGCCUUCCAGC .((..(((.(((((........((((..((((((.((....)).)))))).))))))))))))..))...((((((......(((((.(..(((((....)))))..)))))).)))))) (-46.09 = -46.27 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:12 2006