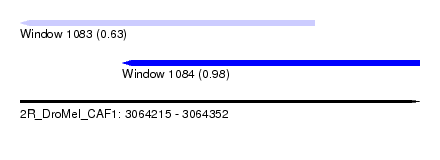
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,064,215 – 3,064,352 |
| Length | 137 |
| Max. P | 0.975892 |

| Location | 3,064,215 – 3,064,316 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.18 |
| Mean single sequence MFE | -20.73 |
| Consensus MFE | -12.51 |
| Energy contribution | -13.45 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
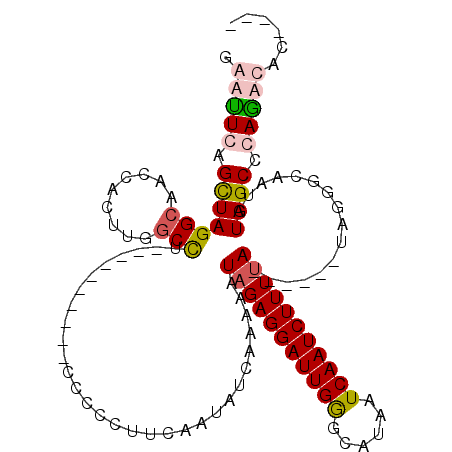
>2R_DroMel_CAF1 3064215 101 - 20766785 CUUUUAAUAUCUUAAAUAGAGGAUUGAGCAUAAUCAAUCUUUUA------UAGGGCAAUGUAGCCCAAACAA----ACACAUUCUGAAUUGAAAGGAAAUCAUUAAAAAUG ((((((((.((....((((((((((((......)))))))))))------).((((......))))......----.........))))))))))................ ( -25.70) >DroSim_CAF1 15510 105 - 1 CAUUCCAUAUCUUAAAUAGAGGAUUGGGCAUAAUCAAUCUUUUA------UGGGGCAAUGUAGCCCAGACACACACACACACUCUGAAUUGAAAGGAAAUCAUUAAAAAUG ..((((.........((((((((((((......)))))))))))------).((((......))))............................))))............. ( -23.30) >DroEre_CAF1 15283 100 - 1 UCUUCGAUAUUAAAAAUAGAGGAUUGGGCAUAAUCAAUCUUUCA------AAGGUCUAUGUAGC-CAGGCAC----ACACAUACUGAAUUGAAAGGAAAUCAUUAAAAAUG ..((((((..........(((((((((......)))))))))..------...((((.......-.))))..----...........)))))).................. ( -13.30) >DroYak_CAF1 15363 107 - 1 CCUUUAAUAUCAAAAAUAGAGGAUUGGGCAUAAUCAAUCUUUUAUACUCGUAAGUCUAUGUAGCCCAGGCAC----ACACAUACUAAAUUGAAAGGAAAUCAUUAAAAAGG (((((..........((((((((((((......))))))))))))....(((.((...(((.((....))))----).)).))).......)))))............... ( -20.60) >consensus CCUUCAAUAUCAAAAAUAGAGGAUUGGGCAUAAUCAAUCUUUUA______UAGGGCAAUGUAGCCCAGACAC____ACACAUACUGAAUUGAAAGGAAAUCAUUAAAAAUG ..((((((........(((((((((((......)))))))))))........((((......)))).....................)))))).................. (-12.51 = -13.45 + 0.94)

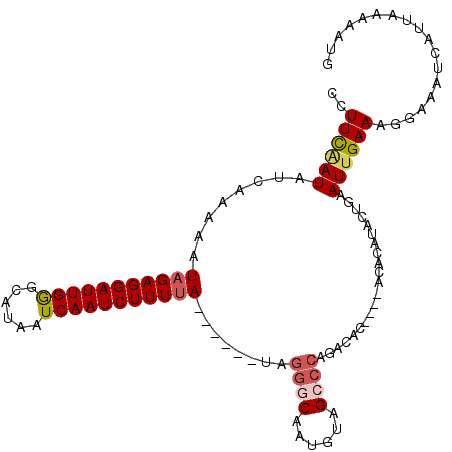
| Location | 3,064,250 – 3,064,352 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.69 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -15.16 |
| Energy contribution | -16.23 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3064250 102 - 20766785 GGUUUGAGCUAGGCAACCAACUGGCUGGG----CUUUCCCCUUUUAAUAUCUUAAAUAGAGGAUUGAGCAUAAUCAAUCUUUUA------UAGGGCAAUGUAGCCCAAACAA---- .((((.((((((........))))))(((----((...((((.((((....))))((((((((((((......)))))))))))------)))))......)))))))))..---- ( -33.90) >DroSim_CAF1 15545 110 - 1 GGUUUGCGUUAGGCAACCGCUUGCCCUGCCCCCCAUUCCCCAUUCCAUAUCUUAAAUAGAGGAUUGGGCAUAAUCAAUCUUUUA------UGGGGCAAUGUAGCCCAGACACACAC .(((((.(((((((((....))))).((((((......((((.(((...(((.....)))))).)))).((((........)))------)))))))...)))).)))))...... ( -34.20) >DroEre_CAF1 15318 95 - 1 GAACUCAGCUAGGCAGCCACUUGGCCU----------CCAUCUUCGAUAUUAAAAAUAGAGGAUUGGGCAUAAUCAAUCUUUCA------AAGGUCUAUGUAGC-CAGGCAC---- .......(((.(((.(((....)))..----------((((((((.............))))).)))(((((....((((....------.)))).))))).))-).)))..---- ( -23.42) >DroYak_CAF1 15398 102 - 1 GAAUUCAGCUAGGCAACCACUUGGCCU----------CCCCCUUUAAUAUCAAAAAUAGAGGAUUGGGCAUAAUCAAUCUUUUAUACUCGUAAGUCUAUGUAGCCCAGGCAC---- .......(((.(((.........((((----------...((((((..........))))))...)))).............((((..(....)..))))..)))..)))..---- ( -20.20) >consensus GAAUUCAGCUAGGCAACCACUUGGCCU__________CCCCCUUCAAUAUCAAAAAUAGAGGAUUGGGCAUAAUCAAUCUUUUA______UAGGGCAAUGUAGCCCAGACAC____ .(((((.(((((((.........)))..............................(((((((((((......)))))))))))................)))).)))))...... (-15.16 = -16.23 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:31 2006