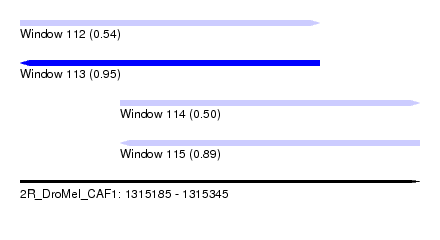
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,315,185 – 1,315,345 |
| Length | 160 |
| Max. P | 0.946711 |

| Location | 1,315,185 – 1,315,305 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.97 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -23.83 |
| Energy contribution | -24.95 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1315185 120 + 20766785 AAUAAAGUCAAUCAAAUAGUGUCUUAAUUGUUAAAGGGCCGUGAAGGGUUUUAUGCCGGAGAAUUGGAUUUAUUCCCAAUGCAAACAUAAACAAUGGGCUGCAAGAUGUCGAGCAGCCAA ....(((.((.........)).))).((((((..((((((......)))))).(((.((.((((.......))))))...)))......)))))).((((((..(....)..)))))).. ( -25.60) >DroSec_CAF1 33580 120 + 1 AACAAAGUCAAUCAAAUAGUGGCUUAAUUGUUAAAGGGCCGUGAAGGGUUUUAUGUCUGCGAAUUGGAUUUAUUCCCAAUACAAACAUAAACAAUGGGCUGCAAGAUGUCGAGCAGCCAA ....((((((.........)))))).((((((...(((((......)))))(((((.((...(((((........))))).)).))))))))))).((((((..(....)..)))))).. ( -29.70) >DroSim_CAF1 12530 120 + 1 AACAAAGUCAAUCAAAUAGUGGCUUAAUCGUUGAAGGGCCGUGAAGGGUUUUAUGUCUGCGAAUUGGAUUUAUUCCCAAUACAAACAUAAACAAUGGGCUGCAAGAUGUCGAGCAGCCAA ....((((((.........))))))...(((((....(((......)))(((((((.((...(((((........))))).)).))))))))))))((((((..(....)..)))))).. ( -30.90) >DroEre_CAF1 16021 120 + 1 AGUAAAGUCAAUCGAAUAGUGGCUAAAUUGUUAAAAAGCCAUAAAGGGUUUUAUGUCUGAAAAUUGGAUUUACUCCCAAUACAAACAUUAACAAUGGGCUGGAAGUCGUCGAGCAGCCAA ...................(((((..((((((((((((((......)))))).(((.((...(((((........))))).)).)))))))))))..(((.((.....)).)))))))). ( -28.20) >consensus AACAAAGUCAAUCAAAUAGUGGCUUAAUUGUUAAAGGGCCGUGAAGGGUUUUAUGUCUGAGAAUUGGAUUUAUUCCCAAUACAAACAUAAACAAUGGGCUGCAAGAUGUCGAGCAGCCAA ....((((((.........)))))).((((((..((((((......))))))((((.((...(((((........))))).)).)))).)))))).((((((..(....)..)))))).. (-23.83 = -24.95 + 1.12)

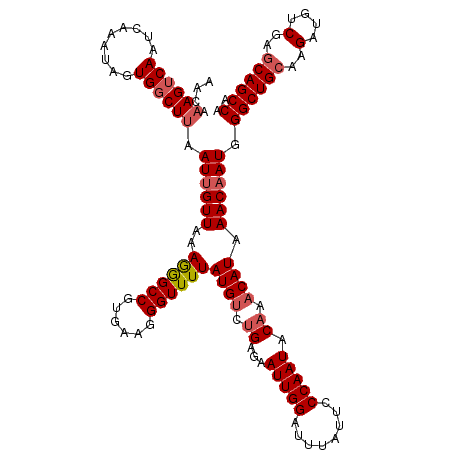
| Location | 1,315,185 – 1,315,305 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.97 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -24.48 |
| Energy contribution | -25.10 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.946711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1315185 120 - 20766785 UUGGCUGCUCGACAUCUUGCAGCCCAUUGUUUAUGUUUGCAUUGGGAAUAAAUCCAAUUCUCCGGCAUAAAACCCUUCACGGCCCUUUAACAAUUAAGACACUAUUUGAUUGACUUUAUU ..((((((..(....)..)))))).....((((((((.(.((((((......)))))).)...))))))))...................((((((((......))))))))........ ( -24.30) >DroSec_CAF1 33580 120 - 1 UUGGCUGCUCGACAUCUUGCAGCCCAUUGUUUAUGUUUGUAUUGGGAAUAAAUCCAAUUCGCAGACAUAAAACCCUUCACGGCCCUUUAACAAUUAAGCCACUAUUUGAUUGACUUUGUU ..((((((..(....)..)))))).....(((((((((((((((((......))))))..)))))))))))...................((((((((......))))))))........ ( -30.30) >DroSim_CAF1 12530 120 - 1 UUGGCUGCUCGACAUCUUGCAGCCCAUUGUUUAUGUUUGUAUUGGGAAUAAAUCCAAUUCGCAGACAUAAAACCCUUCACGGCCCUUCAACGAUUAAGCCACUAUUUGAUUGACUUUGUU ..((((((..(....)..)))))).....(((((((((((((((((......))))))..)))))))))))...................((((((((......))))))))........ ( -30.00) >DroEre_CAF1 16021 120 - 1 UUGGCUGCUCGACGACUUCCAGCCCAUUGUUAAUGUUUGUAUUGGGAGUAAAUCCAAUUUUCAGACAUAAAACCCUUUAUGGCUUUUUAACAAUUUAGCCACUAUUCGAUUGACUUUACU .((((((((.((.....)).)))..((((((((((((((.((((((......))))))...)))))))((((.((.....)).)))))))))))..)))))................... ( -25.70) >consensus UUGGCUGCUCGACAUCUUGCAGCCCAUUGUUUAUGUUUGUAUUGGGAAUAAAUCCAAUUCGCAGACAUAAAACCCUUCACGGCCCUUUAACAAUUAAGCCACUAUUUGAUUGACUUUAUU ..((((((..(....)..)))))).....((((((((((.((((((......))))))...))))))))))...................((((((((......))))))))........ (-24.48 = -25.10 + 0.62)

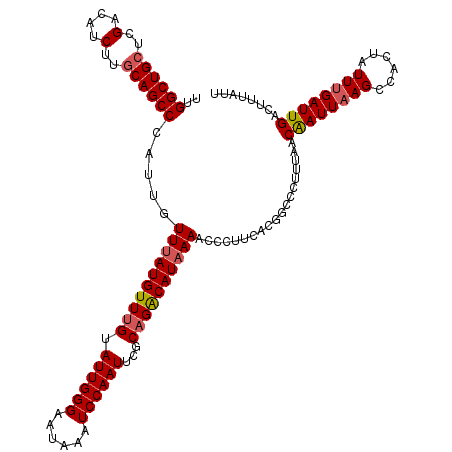
| Location | 1,315,225 – 1,315,345 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -23.34 |
| Energy contribution | -24.27 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1315225 120 + 20766785 GUGAAGGGUUUUAUGCCGGAGAAUUGGAUUUAUUCCCAAUGCAAACAUAAACAAUGGGCUGCAAGAUGUCGAGCAGCCAAUUCAAAUUAGAUCCCCUGCCUAAAGCUUCACAAAUUAAGC ((((..(((((((.((.((.(((((((........)))))................((((((..(....)..)))))).............)).)).)).)))))))))))......... ( -32.20) >DroSec_CAF1 33620 120 + 1 GUGAAGGGUUUUAUGUCUGCGAAUUGGAUUUAUUCCCAAUACAAACAUAAACAAUGGGCUGCAAGAUGUCGAGCAGCCAAUUCAAAUUAAAUCCCCUGCCGGAGGCUUUACAAAUUAAGC ((((((((.(((((((.((...(((((........))))).)).))))))).....((((((..(....)..))))))...............))))(((...))).))))......... ( -31.00) >DroSim_CAF1 12570 120 + 1 GUGAAGGGUUUUAUGUCUGCGAAUUGGAUUUAUUCCCAAUACAAACAUAAACAAUGGGCUGCAAGAUGUCGAGCAGCCAAUUCAAAUUAGAUCCCCUGCCGGAGGCUUUACAAAUUAAGC ((((((((.(((((((.((...(((((........))))).)).))))))).....((((((..(....)..))))))...............))))(((...))).))))......... ( -31.00) >DroEre_CAF1 16061 120 + 1 AUAAAGGGUUUUAUGUCUGAAAAUUGGAUUUACUCCCAAUACAAACAUUAACAAUGGGCUGGAAGUCGUCGAGCAGCCAAUUCAAAUUACAACUCCUUCCUGGAGCUUUACAAACGGAUC .(((((.(((..((((.((...(((((........))))).)).)))).)))....(((((...(....)...)))))..............((((.....))))))))).......... ( -22.50) >consensus GUGAAGGGUUUUAUGUCUGAGAAUUGGAUUUAUUCCCAAUACAAACAUAAACAAUGGGCUGCAAGAUGUCGAGCAGCCAAUUCAAAUUAGAUCCCCUGCCGGAAGCUUUACAAAUUAAGC ((((((((.(((((((.((...(((((........))))).)).))))))).....((((((..(....)..))))))...............))))((.....)).))))......... (-23.34 = -24.27 + 0.94)

| Location | 1,315,225 – 1,315,345 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.27 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1315225 120 - 20766785 GCUUAAUUUGUGAAGCUUUAGGCAGGGGAUCUAAUUUGAAUUGGCUGCUCGACAUCUUGCAGCCCAUUGUUUAUGUUUGCAUUGGGAAUAAAUCCAAUUCUCCGGCAUAAAACCCUUCAC ((((((...(.....).))))))(((((..........(((.((((((..(....)..)))))).))).((((((((.(.((((((......)))))).)...)))))))).)))))... ( -31.90) >DroSec_CAF1 33620 120 - 1 GCUUAAUUUGUAAAGCCUCCGGCAGGGGAUUUAAUUUGAAUUGGCUGCUCGACAUCUUGCAGCCCAUUGUUUAUGUUUGUAUUGGGAAUAAAUCCAAUUCGCAGACAUAAAACCCUUCAC ..............(((...)))(((((..........(((.((((((..(....)..)))))).))).(((((((((((((((((......))))))..))))))))))).)))))... ( -35.90) >DroSim_CAF1 12570 120 - 1 GCUUAAUUUGUAAAGCCUCCGGCAGGGGAUCUAAUUUGAAUUGGCUGCUCGACAUCUUGCAGCCCAUUGUUUAUGUUUGUAUUGGGAAUAAAUCCAAUUCGCAGACAUAAAACCCUUCAC ..............(((...)))(((((..........(((.((((((..(....)..)))))).))).(((((((((((((((((......))))))..))))))))))).)))))... ( -35.90) >DroEre_CAF1 16061 120 - 1 GAUCCGUUUGUAAAGCUCCAGGAAGGAGUUGUAAUUUGAAUUGGCUGCUCGACGACUUCCAGCCCAUUGUUAAUGUUUGUAUUGGGAGUAAAUCCAAUUUUCAGACAUAAAACCCUUUAU .....((((....((((((.....))))))........(((.(((((............))))).)))....(((((((.((((((......))))))...))))))).))))....... ( -25.50) >consensus GCUUAAUUUGUAAAGCCUCAGGCAGGGGAUCUAAUUUGAAUUGGCUGCUCGACAUCUUGCAGCCCAUUGUUUAUGUUUGUAUUGGGAAUAAAUCCAAUUCGCAGACAUAAAACCCUUCAC .........((((((((((.....))))..........(((.((((((..(....)..)))))).))).((((((((((.((((((......))))))...))))))))))...)))))) (-27.34 = -27.27 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:25:09 2006